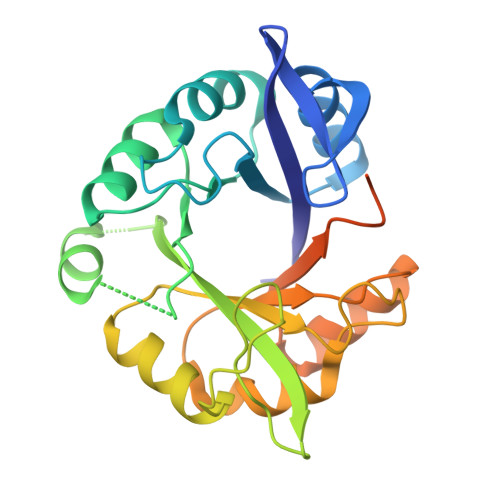
High-Resolution Crystal Structure of an Artificial (Betaalpha)(8)-Barrel Protein Designed from Identical Half-Barrels.
Hocker, B., Lochner, A., Seitz, T., Claren, J., Sterner, R.(2009) Biochemistry 48: 1145
- PubMed: 19166324
- DOI: https://doi.org/10.1021/bi802125b
- Primary Citation of Related Structures:
2W6R - PubMed Abstract:
Ample evidence suggests that the ubiquitous (betaalpha)(8)-barrel enzyme fold has evolved by the duplication and fusion of an ancestral (betaalpha)(4)-half-barrel. To reconstruct this process in the laboratory with a model protein, we earlier fused two copies of the C-terminal half-barrel HisF-C of imidazole glycerol phosphate synthase (HisF) and stepwise stabilized the resulting HisF-CC construct. We now further increased its stability and solubility by introducing two additional amino acid exchanges, which allowed us to crystallize the resulting artificial (betaalpha)(8)-barrel protein HisF-C***C. The analysis of its X-ray structure at 2.1 A resolution reveals a striking similarity to wild-type HisF, helps us to understand its improved stability, and provides further insights into the evolution of (betaalpha)(8)-barrel proteins.
Organizational Affiliation:
Max Planck Institute for Developmental Biology, Spemannstrasse 35, D-72076 Tubingen, Germany. birte.hoecker@tuebingen.mpg.de