Cellular Inhibition of Chk2 Kinase and Potentiation of Camptothecins and Radiation by the Novel Chk2 Inhibitor Pv1019.
Jobson, A.G., Lountos, G.T., Lorenzi, P.L., Llamas, J., Connelly, J., Cerna, D., Tropea, J.E., Onda, A., Zoppoli, G., Kondapaka, S., Zhang, G., Caplen, N.J., Cardellina, J.H., Yoo, S.S., Monks, A., Self, C., Waugh, D.S., Shoemaker, R.H., Pommier, Y.(2009) J Pharmacol Exp Ther 331: 816
- PubMed: 19741151
- DOI: https://doi.org/10.1124/jpet.109.154997
- Primary Citation of Related Structures:
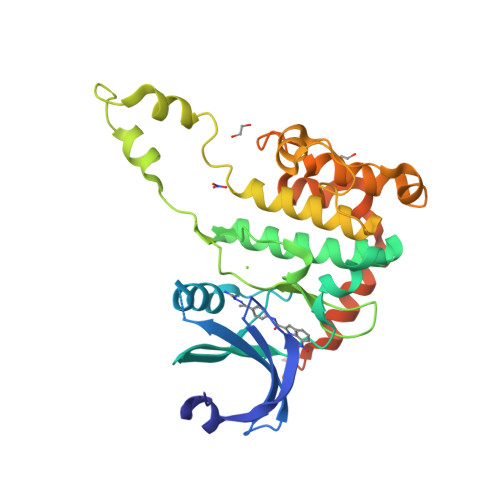
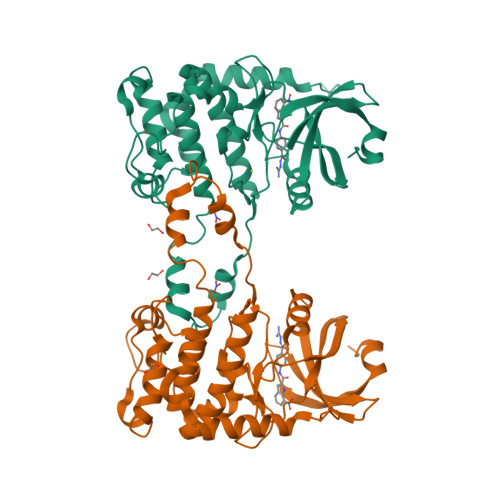
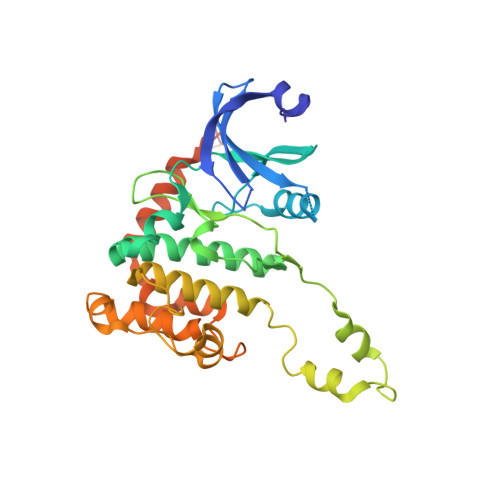
2W7X - PubMed Abstract:
Chk2 is a checkpoint kinase involved in the ataxia telangiectasia mutated pathway, which is activated by genomic instability and DNA damage, leading to either cell death (apoptosis) or cell cycle arrest. Chk2 provides an unexplored therapeutic target against cancer cells. We recently reported 4,4'-diacetyldiphenylurea-bis(guanylhydrazone) (NSC 109555) as a novel chemotype Chk2 inhibitor. We have now synthesized a derivative of NSC 109555, PV1019 (NSC 744039) [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide], which is a selective submicromolar inhibitor of Chk2 in vitro. The cocrystal structure of PV1019 bound in the ATP binding pocket of Chk2 confirmed enzymatic/biochemical observations that PV1019 acts as a competitive inhibitor of Chk2 with respect to ATP. PV1019 was found to inhibit Chk2 in cells. It inhibits Chk2 autophosphorylation (which represents the cellular kinase activation of Chk2), Cdc25C phosphorylation, and HDMX degradation in response to DNA damage. PV1019 also protects normal mouse thymocytes against ionizing radiation-induced apoptosis, and it shows synergistic antiproliferative activity with topotecan, camptothecin, and radiation in human tumor cell lines. We also show that PV1019 and Chk2 small interfering RNAs can exert antiproliferative activity themselves in the cancer cells with high Chk2 expression in the NCI-60 screen. These data indicate that PV1019 is a potent and selective inhibitor of Chk2 with chemotherapeutic and radiosensitization potential.
Organizational Affiliation:
Laboratory of Molecular Pharmacology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892-4255, USA.