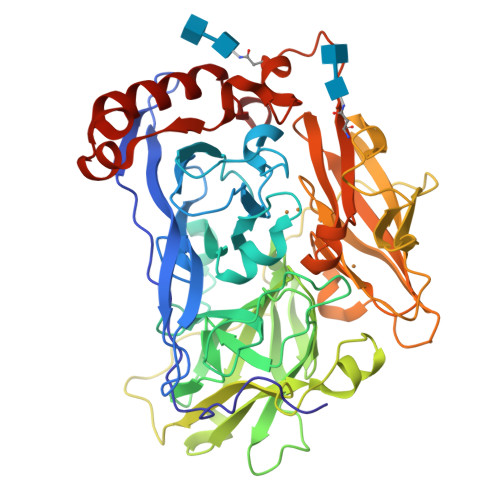
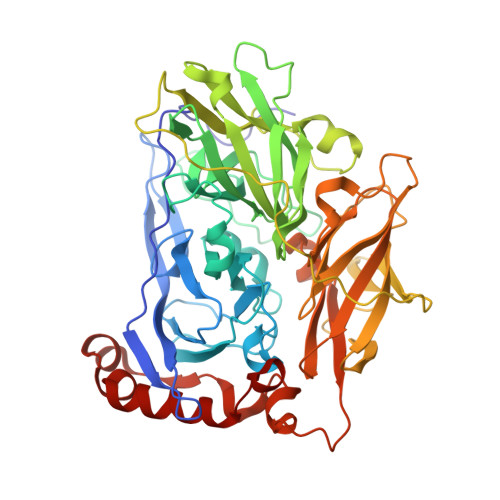
Bilirubin Oxidase from Myrothecium Verrucaria: X- Ray Determination of the Complete Crystal Structure and a Rational Surface Modification for Enhanced Electrocatalytic O(2) Reduction.
Cracknell, J.A., Mcnamara, T.P., Lowe, E.D., Blanford, C.F.(2011) Dalton Trans 40: 6668
- PubMed: 21544308
- DOI: https://doi.org/10.1039/c0dt01403f
- Primary Citation of Related Structures:
2XLL - PubMed Abstract:
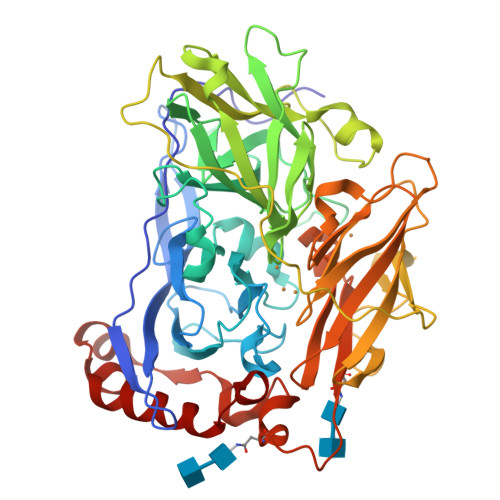
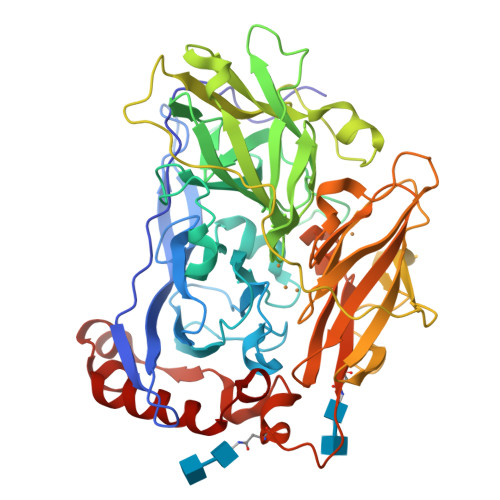
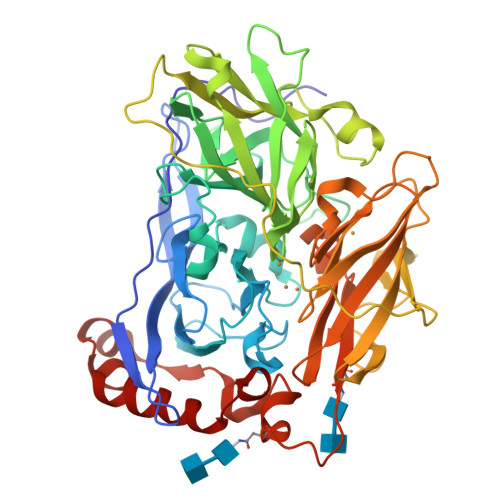
The blue multi-copper oxidase bilirubin oxidase (BOx) from the ascomycete plant pathogen Myrothecium verrucaria (Mv) efficiently catalyses the oxidation of bilirubin to biliverdin, with the concomitant reduction of O(2) to water, a reaction of considerable interest for low-temperature bio-fuel cell applications. We have solved the complete X-ray determined structure of Mv BOx at 2.4 Å resolution, using molecular replacement with the Spore Coat Protein A (CotA) enzyme from Bacillus subtilis (PDB code 1GSK) as a template. The structure reveals an unusual environment around the blue type 1 copper (T1 Cu) that includes two non-coordinating hydrophilic amino acids, asparagine and threonine. The presence of a long, narrow and hydrophilic pocket near the T1 Cu suggests that structure of the substrate-binding site is dynamically determined in vivo. We show that the interaction between the binding pocket of Mv BOx and its highly conjugated natural organic substrate, bilirubin, can be used to stabilise the enzyme on a pyrolytic graphite electrode, more than doubling its electrocatalytic activity relative to the current obtained by simple adsorption of the protein to the carbon surface.
Organizational Affiliation:
Department of Chemistry, Inorganic Chemistry Laboratory, University of Oxford, South Parks Road, Oxford, UK OX1 3QR.