Structural Basis for Modification of Flavonol and Naphthol Glucoconjugates by Nicotiana Tabacum Malonyltransferase (Ntmat1).
Manjasetty, B.A., Yu, X.H., Panjikar, S., Taguchi, G., Chance, M.R., Li, C.J.(2012) Planta 236: 781
- PubMed: 22610270
- DOI: https://doi.org/10.1007/s00425-012-1660-8
- Primary Citation of Related Structures:
2XR7 - PubMed Abstract:
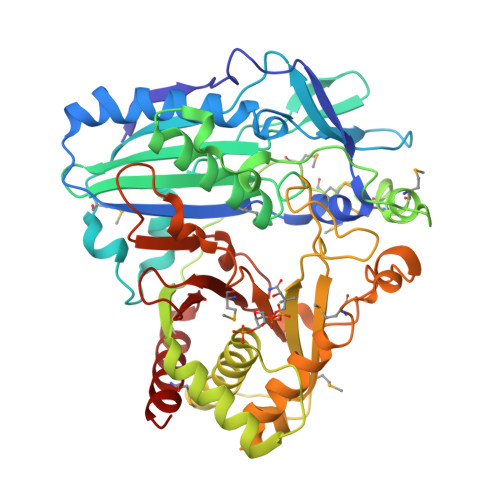
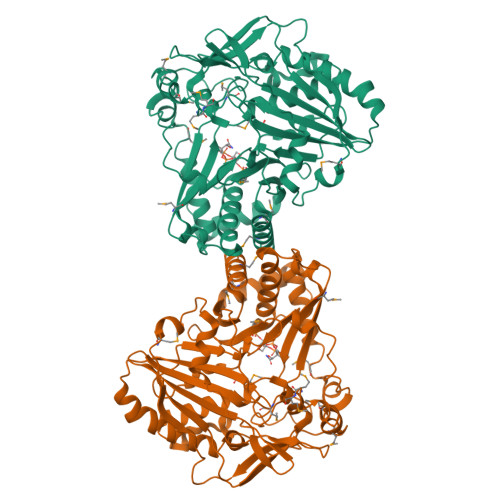
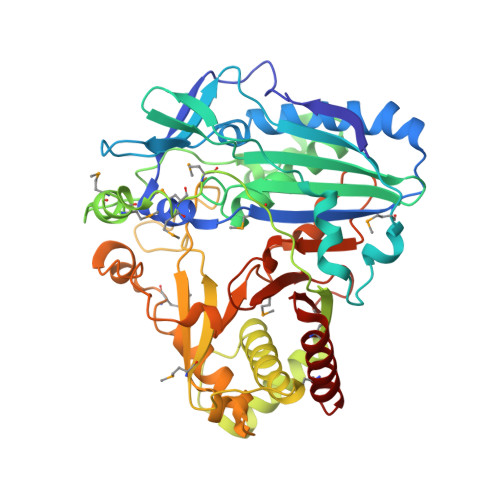
Plant HXXXD acyltransferase-catalyzed malonylation is an important modification reaction in elaborating the structural diversity of flavonoids and anthocyanins, and a universal adaptive mechanism to detoxify xenobiotics. Nicotiana tabacum malonyltransferase 1 (NtMaT1) is a member of anthocyanin acyltransferase subfamily that uses malonyl-CoA (MLC) as donor catalyzing transacylation in a range of flavonoid and naphthol glucosides. To gain insights into the molecular basis underlying its catalytic mechanism and versatile substrate specificity, we resolved the X-ray crystal structure of NtMaT1 to 3.1 Å resolution. The structure comprises two α/β mixed subdomains, as typically found in the HXXXD acyltransferases. The partial electron density map of malonyl-CoA allowed us to reliably dock the entire molecule into the solvent channel and subsequently define the binding sites for both donor and acceptor substrates. MLC bound to the NtMaT1 occupies one end of the long solvent channel between two subdomains. On superimposing and comparing the structure of NtMaT1 with that of an enzyme from anthocyanin acyltransferase subfamily from red chrysanthemum (Dm3Mat3) revealed large architectural variation in the binding sites, both for the acyl donor and for the acceptor, although their overall protein folds are structurally conserved. Consequently, the shape and the interactions of malonyl-CoA with the binding sites' amino acid residues differ substantially. These major local architectural disparities point to the independent, divergent evolution of plant HXXXD acyltransferases in different species. The structural flexibility of the enzyme and the amendable binding pattern of the substrates provide a basis for the evolution of the distinct, versatile substrate specificity of plant HXXXD acyltransferases.
Organizational Affiliation:
European Molecular Biology Laboratory, Grenoble Outstation and Unit of Virus Host-Cell Interactions, UJF-EMBL-CNRS, UMI 3265, 6 Rue Jules Horowitz, 38042, Grenoble Cedex 9, France.