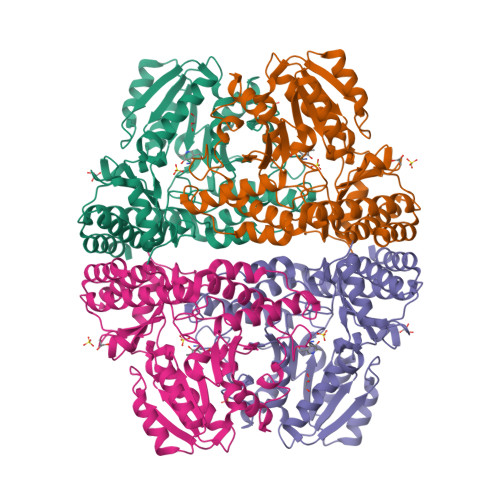
Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
Yoshikane, Y., Yokochi, N., Yamasaki, M., Mizutani, K., Ohnishi, K., Mikami, B., Hayashi, H., Yagi, T.(2008) J Biological Chem 283: 1120-1127
- PubMed: 17989071
- DOI: https://doi.org/10.1074/jbc.M708061200
- Primary Citation of Related Structures:
2Z9U, 2Z9V, 2Z9W, 2Z9X - PubMed Abstract:
Pyridoxamine-pyruvate aminotransferase (PPAT; EC 2.6.1.30) is a pyridoxal 5'-phosphate-independent aminotransferase and catalyzes reversible transamination between pyridoxamine and pyruvate to form pyridoxal and L-alanine. The crystal structure of PPAT from Mesorhizobium loti has been solved in space group P4(3)2(1)2 and was refined to an R factor of 15.6% (R(free) = 20.6%) at 2.0 A resolution. In addition, the structures of PPAT in complexes with pyridoxamine, pyridoxal, and pyridoxyl-L-alanine have been refined to R factors of 15.6, 15.4, and 14.5% (R(free) = 18.6, 18.1, and 18.4%) at 1.7, 1.7, and 2.0 A resolution, respectively. PPAT is a homotetramer and each subunit is composed of a large N-terminal domain, consisting of seven beta-sheets and eight alpha-helices, and a smaller C-terminal domain, consisting of three beta-sheets and four alpha-helices. The substrate pyridoxal is bound through an aldimine linkage to Lys-197 in the active site. The alpha-carboxylate group of the substrate amino/keto acid is hydrogen-bonded to Arg-336 and Arg-345. The structures revealed that the bulky side chain of Glu-68 interfered with the binding of the phosphate moiety of pyridoxal 5'-phosphate and made PPAT specific to pyridoxal. The reaction mechanism of the enzyme is discussed based on the structures and kinetics results.
Organizational Affiliation:
Department of Bioresources Science, Faculty of Agriculture, Kochi University, Monobe-Otsu 200, Nankoku, Kochi 783-8502, Japan.