Rational Design of a beta-Lactamase Inhibitor Achieved via Stabilization of the trans-Enamine Intermediate: 1.28 A Crystal Structure of wt SHV-1 Complex with a Penam Sulfone.
Padayatti, P.S., Sheri, A., Totir, M.A., Helfand, M.S., Carey, M.P., Anderson, V.E., Carey, P.R., Bethel, C.R., Bonomo, R.A., Buynak, J.D., van den Akker, F.(2006) J Am Chem Soc 128: 13235-13242
- PubMed: 17017804
- DOI: https://doi.org/10.1021/ja063715w
- Primary Citation of Related Structures:
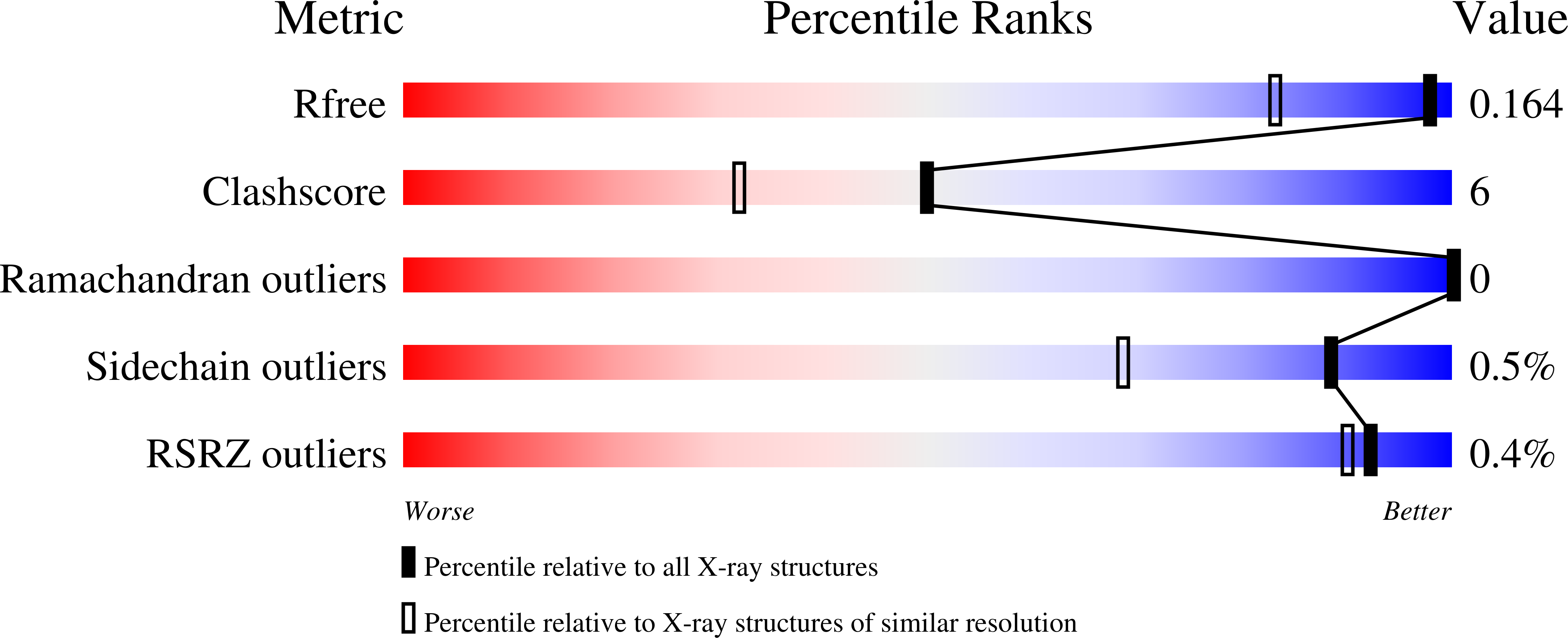
2H5S - PubMed Abstract:
beta-Lactamases are one of the major causes of antibiotic resistance in Gram negative bacteria. The continuing evolution of beta-lactamases that are capable of hydrolyzing our most potent beta-lactams presents a vexing clinical problem, in particular since a number of them are resistant to inhibitors. The efficient inhibition of these enzymes is therefore of great clinical importance. Building upon our previous structural studies that examined tazobactam trapped as a trans-enamine intermediate in a deacylation deficient SHV variant, we designed a novel penam sulfone derivative that forms a more stable trans-enamine intermediate. We report here the 1.28 A resolution crystal structure of wt SHV-1 in complex with a rationally designed penam sulfone, SA2-13. The compound is covalently bound to the active site of wt SHV-1 similar to tazobactam yet forms an additional salt-bridge with K234 and hydrogen bonds with S130 and T235 to stabilize the trans-enamine intermediate. Kinetic measurements show that SA2-13, once reacted with SHV-1 beta-lactamase, is about 10-fold slower at being released from the enzyme compared to tazobactam. Stabilizing the trans-enamine intermediate represents a novel strategy for the rational design of mechanism-based class A beta-lactamase inhibitors.
Organizational Affiliation:
Department of Biochemistry, Case Western Reserve University, Cleveland, Ohio 44106, USA.