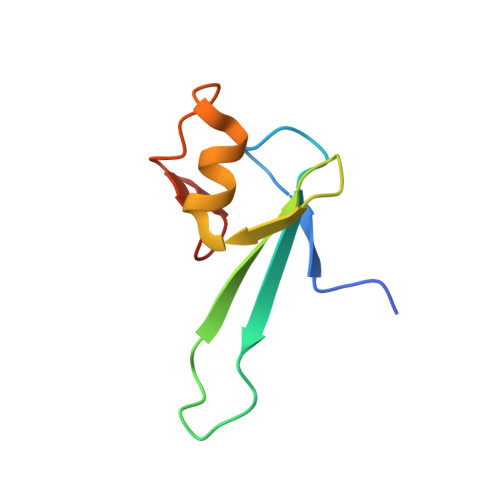
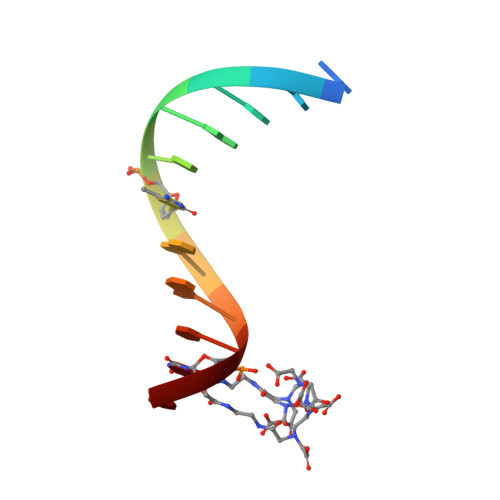
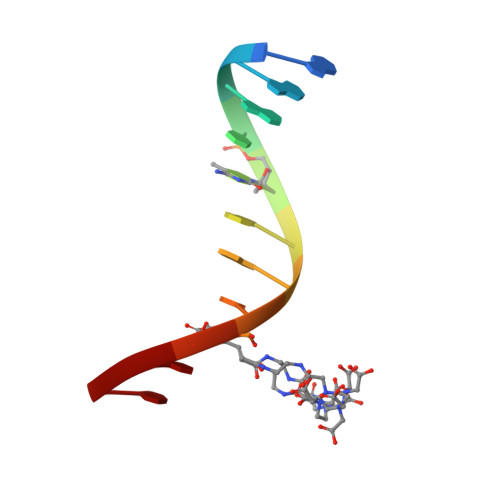
Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence.
Scarsdale, J.N., Webb, H.D., Ginder, G.D., Williams, D.C.(2011) Nucleic Acids Res 39: 6741-6752
- PubMed: 21531701
- DOI: https://doi.org/10.1093/nar/gkr262
- Primary Citation of Related Structures:
2KY8 - PubMed Abstract:
The epigenetic code of DNA methylation is interpreted chiefly by methyl cytosine binding domain (MBD) proteins which in turn recruit multiprotein co-repressor complexes. We previously isolated one such complex, MBD2-NuRD, from primary erythroid cells and have shown it contributes to embryonic/fetal β-type globin gene silencing during development. This complex has been implicated in silencing tumor suppressor genes in a variety of human tumor cell types. Here we present structural details of chicken MBD2 bound to a methylated DNA sequence from the ρ-globin promoter to which it binds in vivo and mediates developmental transcriptional silencing in normal erythroid cells. While previous studies have failed to show sequence specificity for MBD2 outside of the symmetric mCpG, we find that this domain binds in a single orientation on the ρ-globin target DNA sequence. Further, we show that the orientation and affinity depends on guanine immediately following the mCpG dinucleotide. Dynamic analyses show that DNA binding stabilizes the central β-sheet, while the N- and C-terminal regions of the protein maintain mobility. Taken together, these data lead to a model in which DNA binding stabilizes the MBD2 structure and that binding orientation and affinity is influenced by the DNA sequence surrounding the central mCpG.
Organizational Affiliation:
Institute of Structural Biology and Drug Design, Virginia Commonwealth University, Richmond, VA 23298-0035, USA.