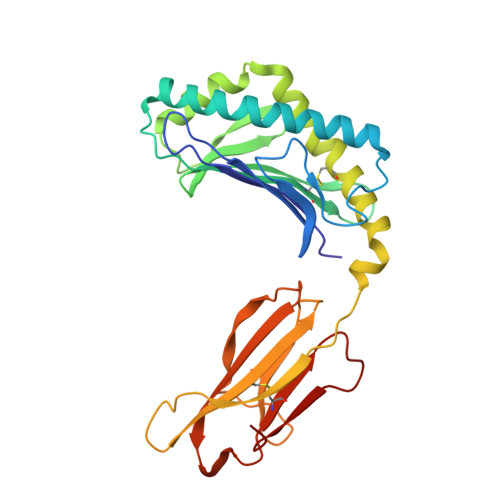
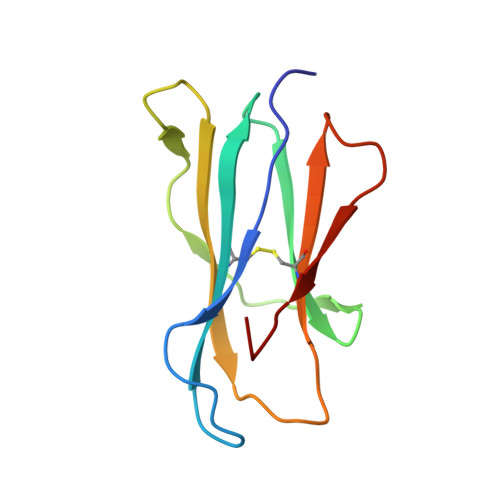
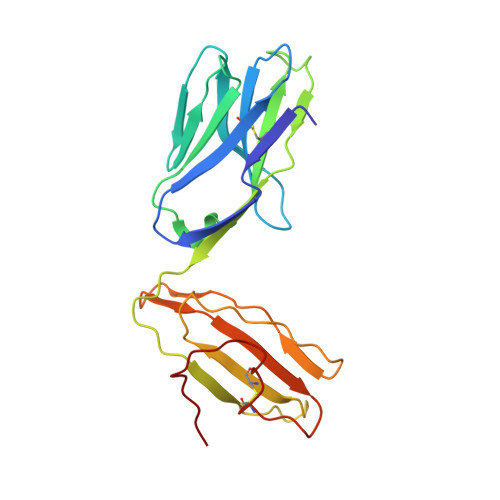
CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor.
Borg, N.A., Wun, K.S., Kjer-Nielsen, L., Wilce, M.C., Pellicci, D.G., Koh, R., Besra, G.S., Bharadwaj, M., Godfrey, D.I., McCluskey, J., Rossjohn, J.(2007) Nature 448: 44-49
- PubMed: 17581592
- DOI: https://doi.org/10.1038/nature05907
- Primary Citation of Related Structures:
2PO6 - PubMed Abstract:
The CD1 family is a large cluster of non-polymorphic, major histocompatibility complex (MHC) class-I-like molecules that bind distinct lipid-based antigens that are recognized by T cells. The most studied group of T cells that interact with lipid antigens are natural killer T (NKT) cells, which characteristically express a semi-invariant T-cell receptor (NKT TCR) that specifically recognizes the CD1 family member, CD1d. NKT-cell-mediated recognition of the CD1d-antigen complex has been implicated in microbial immunity, tumour immunity, autoimmunity and allergy. Here we describe the structure of a human NKT TCR in complex with CD1d bound to the potent NKT-cell agonist alpha-galactosylceramide, the archetypal CD1d-restricted glycolipid. In contrast to T-cell receptor-peptide-antigen-MHC complexes, the NKT TCR docked parallel to, and at the extreme end of the CD1d-binding cleft, which enables a lock-and-key type interaction with the lipid antigen. The structure provides a basis for the interaction between the highly conserved NKT TCR alpha-chain and the CD1d-antigen complex that is typified in innate immunity, and also indicates how variability of the NKT TCR beta-chain can impact on recognition of other CD1d-antigen complexes. These findings provide direct insight into how a T-cell receptor recognizes a lipid-antigen-presenting molecule of the immune system.
Organizational Affiliation:
The Protein Crystallography Unit, ARC Centre of Excellence in Structural and Functional Microbial Genomics, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.