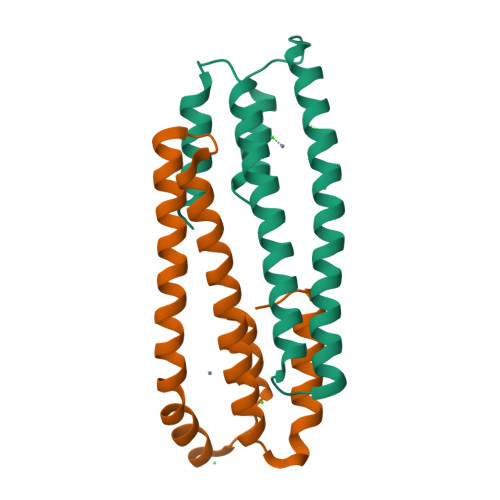
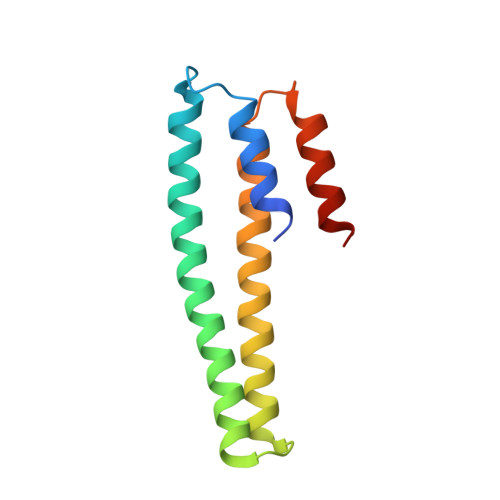
Structural Basis for Metal Sensing by Cnrx.
Trepreau, J., Girard, E., Maillard, A.P., De Rosny, E., Petit-Haertlein, I., Kahn, R., Coves, J.(2011) J Mol Biology 408: 766
- PubMed: 21414325
- DOI: https://doi.org/10.1016/j.jmb.2011.03.014
- Primary Citation of Related Structures:
2Y39, 2Y3B, 2Y3D, 2Y3G, 2Y3H - PubMed Abstract:
CnrX is the metal sensor and signal modulator of the three-protein transmembrane signal transduction complex CnrYXH of Cupriavidus metallidurans CH34 that is involved in the setup of cobalt and nickel resistance. We have determined the atomic structure of the soluble domain of CnrX in its Ni-bound, Co-bound, or Zn-bound form. Ni and Co ions elicit a biological response, while the Zn-bound form is inactive. The structures presented here reveal the topology of intraprotomer and interprotomer interactions and the ability of metal-binding sites to fine-tune the packing of CnrX dimer as a function of the bound metal. These data suggest an allosteric mechanism to explain how the complex is switched on and how the signal is modulated by Ni or Co binding. These results provide clues to propose a model for signal propagation through the membrane in the complex.
Organizational Affiliation:
Institut de Biologie Structurale Jean-Pierre Ebel, UMR 5075, CNRS-CEA-UJF Grenoble 1, 41, rue Jules Horowitz, 38027 Grenoble Cedex, France.