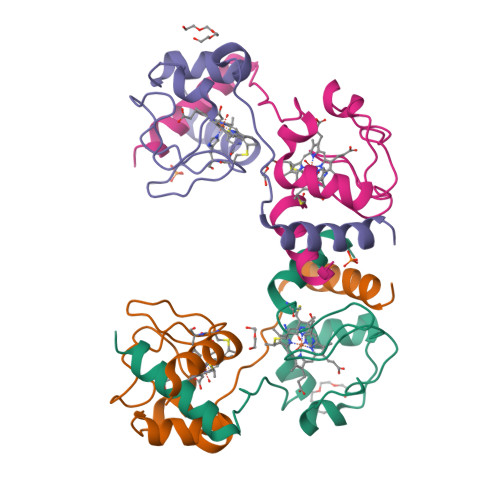
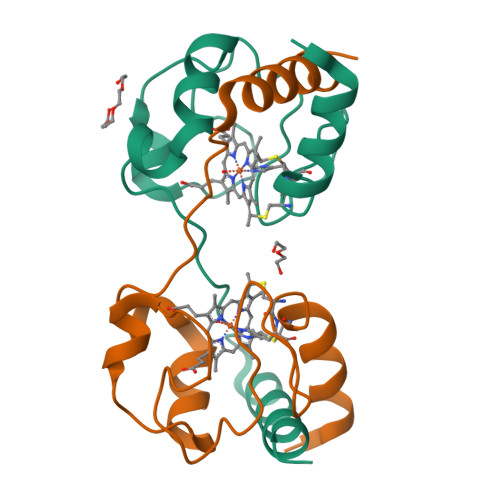
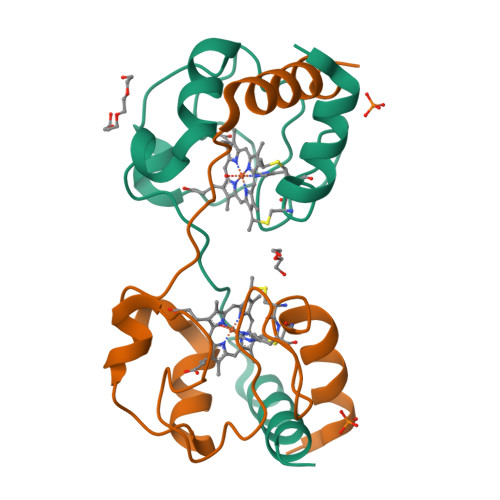
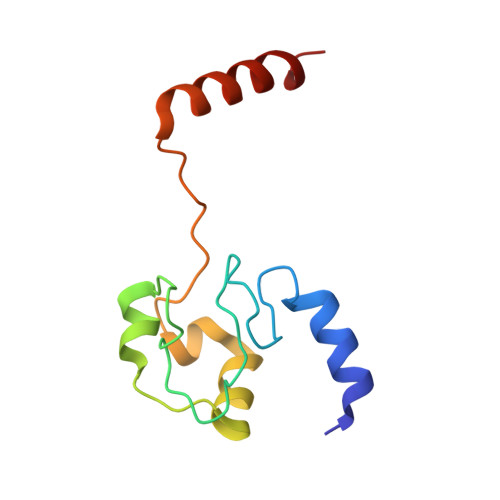
Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Hirota, S., Hattori, Y., Nagao, S., Taketa, M., Komori, H., Kamikubo, H., Wang, Z., Takahashi, I., Negi, S., Sugiura, Y., Kataoka, M., Higuchi, Y.(2010) Proc Natl Acad Sci U S A 107: 12854-12859
- PubMed: 20615990
- DOI: https://doi.org/10.1073/pnas.1001839107
- Primary Citation of Related Structures:
3NBS, 3NBT - PubMed Abstract:
Cytochrome c (cyt c) is a stable protein that functions in a monomeric state as an electron donor for cytochrome c oxidase. It is also released to the cytosol when permeabilization of the mitochondrial outer membrane occurs at the early stage of apoptosis. For nearly half a century, it has been known that cyt c forms polymers, but the polymerization mechanism remains unknown. We found that cyt c forms polymers by successive domain swapping, where the C-terminal helix is displaced from its original position in the monomer and Met-heme coordination is perturbed significantly. In the crystal structures of dimeric and trimeric cyt c, the C-terminal helices are replaced by the corresponding domain of other cyt c molecules and Met80 is dissociated from the heme. The solution structures of dimeric, trimeric, and tetrameric cyt c were linear based on small-angle X-ray scattering measurements, where the trimeric linear structure shifted toward the cyclic structure by addition of PEG and (NH(4))(2)HPO(4). The absorption and CD spectra of high-order oligomers (approximately 40 mer) were similar to those of dimeric and trimeric cyt c but different from those of monomeric cyt c. For dimeric, trimeric, and tetrameric cyt c, the DeltaH of the oligomer dissociation to monomers was estimated to be about -20 kcal/mol per protomer unit, where Met-heme coordination appears to contribute largely to DeltaH. The present results suggest that cyt c polymerization occurs by successive domain swapping, which may be a common mechanism of protein polymerization.
Organizational Affiliation:
Graduate School of Materials Science, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan. hirota@ms.naist.jp