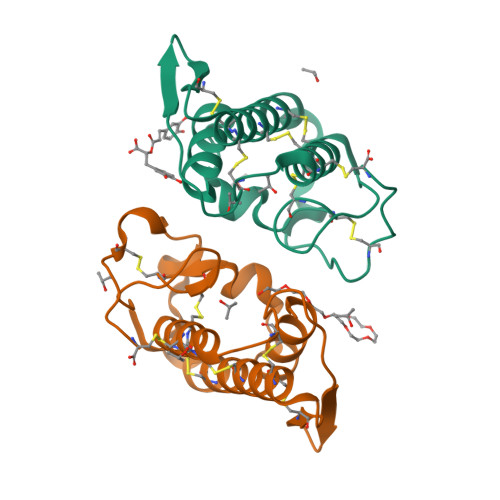
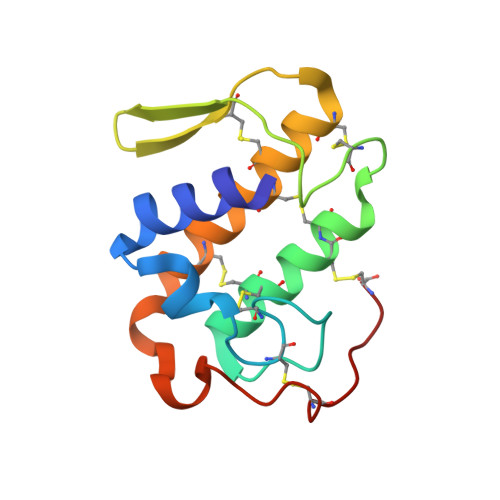
Structural and Functional Studies of a Bothropic Myotoxin Complexed to Rosmarinic Acid: New Insights into Lys49-PLA(2) Inhibition.
Dos Santos, J.I., Cardoso, F.F., Soares, A.M., Dal Pai Silva, M., Gallacci, M., Fontes, M.R.(2011) PLoS One 6: e28521-e28521
- PubMed: 22205953
- DOI: https://doi.org/10.1371/journal.pone.0028521
- Primary Citation of Related Structures:
3QNL - PubMed Abstract:
Snakebite envenoming is an important public health problem in many tropical and subtropical countries, and is considered a neglected tropical disease by the World Health Organization. Most severe cases are inflicted by species of the families Elapidae and Viperidae, and lead to a number of systemic and local effects in the victim. One of the main problems regarding viperidic accidents is prominent local tissue damage whose pathogenesis is complex and involves the combined actions of a variety of venom components. Phospholipases A₂ (PLA₂s) are the most abundant muscle-damaging components of these venoms. Herein, we report functional and structural studies of PrTX-I, a Lys49-PLA₂ from Bothops pirajai snake venom, and the influence of rosmarinic acid (RA) upon this toxin's activities. RA is a known active component of some plant extracts and has been reported as presenting anti-myotoxic properties related to bothopic envenomation. The myotoxic activity of Lys49-PLA₂s is well established in the literature and although no in vivo neurotoxicity has been observed among these toxins, in vitro neuromuscular blockade has been reported for some of these proteins. Our in vitro studies show that RA drastically reduces both the muscle damage and the neuromuscular blockade exerted by PrTX-I on mice neuromuscular preparations (by ∼80% and ∼90%, respectively). These results support the hypothesis that the two effects are closely related and lead us to suggest that they are consequences of the muscle membrane-destabilizing activity of the Lys49-PLA₂. Although the C-terminal region of these proteins has been reported to comprise the myotoxic site, we demonstrate by X-ray crystallographic studies that RA interacts with PrTX-I in a different region. Consequently, a new mode of Lys49-PLA₂ inhibition is proposed. Comparison of our results with others in the literature suggests possible new ways to inhibit bothropic snake venom myotoxins and improve serum therapy.
Organizational Affiliation:
Departamento de Física e Biofísica, Instituto de Biociências, University Estadual Paulista, Botucatu/Sao Paulo, Brazil.