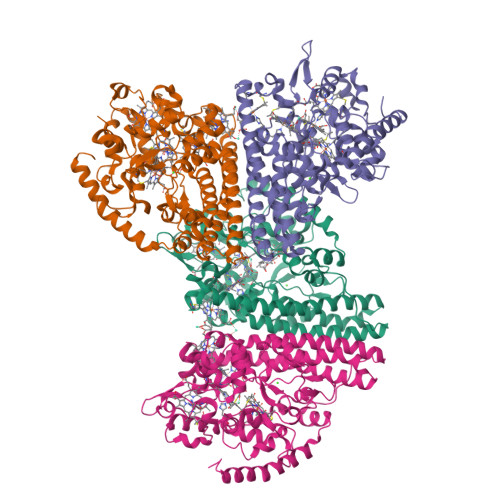
Characterization of the active site and calcium binding in cytochrome c nitrite reductases.
Lockwood, C.W., Clarke, T.A., Butt, J.N., Hemmings, A.M., Richardson, D.J.(2011) Biochem Soc Trans 39: 1871-1875
- PubMed: 22103542
- DOI: https://doi.org/10.1042/BST20110731
- Primary Citation of Related Structures:
3TOR - PubMed Abstract:
The decahaem homodimeric cytochrome c nitrite reductase (NrfA) is expressed within the periplasm of a wide range of Gamma-, Delta- and Epsilon-proteobacteria and is responsible for the six-electron reduction of nitrite to ammonia. This allows nitrite to be used as a terminal electron acceptor, facilitating anaerobic respiration while allowing nitrogen to remain in a biologically available form. NrfA has also been reported to reduce nitric oxide (a reaction intermediate) and sulfite to ammonia and sulfide respectively, suggesting a potential secondary role as a detoxification enzyme. The protein sequences and crystal structures of NrfA from different bacteria and the closely related octahaem nitrite reductase from Thioalkalivibrio nitratireducens (TvNir) reveal that these enzymes are homologous. The NrfA proteins contain five covalently attached haem groups, four of which are bis-histidine-co-ordinated, with the proximal histidine being provided by the highly conserved CXXCH motif. These haems are responsible for intraprotein electron transfer. The remaining haem is the site for nitrite reduction, which is ligated by a novel lysine residue provided by a CXXCK haem-binding motif. The TvNir nitrite reductase has five haems that are structurally similar to those of NrfA and three extra bis-histidine-coordinated haems that precede the NrfA conserved region. The present review compares the protein sequences and structures of NrfA and TvNir and discusses the subtle differences related to active-site architecture and Ca2+ binding that may have an impact on substrate reduction.
Organizational Affiliation:
Centre for Molecular and Structural Biochemistry, School of Chemistry and School of Biological Sciences, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK.