A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Liu, T., Guo, P., Zhou, Y., Wang, J., Chen, L., Yang, H., Qian, X., Yang, Q.(2014) Sci Rep 4: 6188-6188
- PubMed: 25155420
- DOI: https://doi.org/10.1038/srep06188
- Primary Citation of Related Structures:
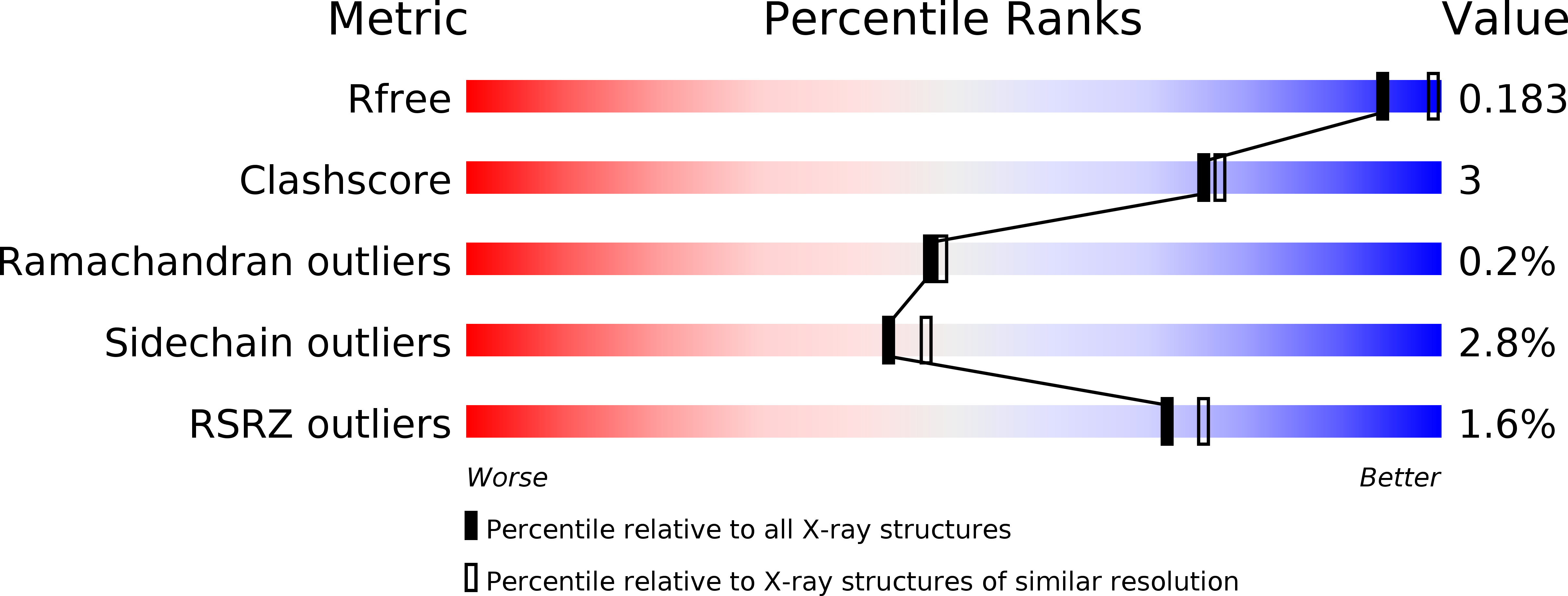
3WMB, 3WMC - PubMed Abstract:
Selective inhibition of function-specific β-GlcNAcase has great potential in terms of drug design and biological research. The symmetrical bis-naphthalimide M-31850 was previously obtained by screening for specificity against human glycoconjugate-lytic β-GlcNAcase. Using protein-ligand co-crystallization and molecular docking, we designed an unsymmetrical dyad of naphthalimide and thiadiazole, Q2, that changes naphthalimide specificity from against a human glycoconjugate-lytic β-GlcNAcase to against insect and bacterial chitinolytic β-GlcNAcases. The crystallographic and in silico studies reveal that the naphthalimide ring can be utilized to bind different parts of these enzyme homologs, providing a new starting point to design specific inhibitors. Moreover, Q2-induced closure of the substrate binding pocket is the structural basis for its 13-fold increment in inhibitory potency. Q2 is the first non-carbohydrate inhibitor against chitinolytic β-GlcNAcases. This study provides a useful example of structure-based rationally designed inhibitors as potential pharmaceuticals or pesticides.
Organizational Affiliation:
1] School of Life Science and Biotechnology, Dalian University of Technology, Dalian 116024, China [2].