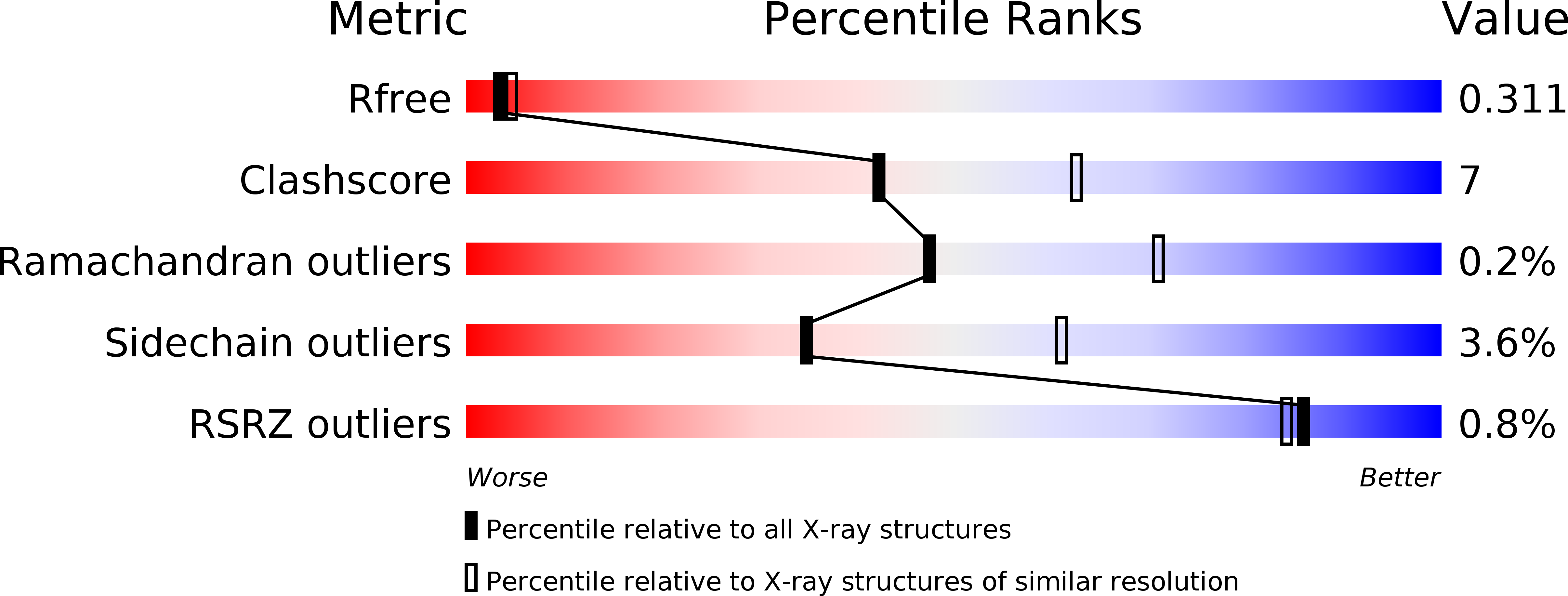
Crystal Structure of Mycobacterium Tuberculosis Zinc-Dependent Metalloprotease-1 (Zmp1), a Metalloprotease Involved in Pathogenicity.
Ferraris, D.M., Sbardella, D., Petrera, A., Marini, S., Amstutz, B., Coletta, M., Sander, P., Rizzi, M.(2011) J Biological Chem 286: 32475
- PubMed: 21813647
- DOI: https://doi.org/10.1074/jbc.M111.271809
- Primary Citation of Related Structures:
3ZUK - PubMed Abstract:
Mycobacterium tuberculosis, the causative agent of tuberculosis, parasitizes host macrophages. The resistance of the tubercle bacilli to the macrophage hostile environment relates to their ability to impair phagosome maturation and its fusion with the lysosome, thus preventing the formation of the phago-lysosome and eventually arresting the process of phagocytosis. The M. tuberculosis zinc-dependent metalloprotease Zmp1 has been proposed to play a key role in the process of phagosome maturation inhibition and emerged as an important player in pathogenesis. Here, we report the crystal structure of wild-type Zmp1 at 2.6 Å resolution in complex with the generic zinc metalloprotease inhibitor phosphoramidon, which we demonstrated to inhibit the enzyme potently. Our data represent the first structural characterization of a bacterial member of the zinc-dependent M13 endopeptidase family and revealed a significant degree of conservation with eukaryotic enzymes. However, structural comparison of the Zmp1-phosphoramidon complex with homologous human proteins neprilysin and endothelin-converting enzyme-1 revealed unique features of the Zmp1 active site to be exploited for the rational design of specific inhibitors that may prove useful as a pharmacological tool for better understanding Zmp1 biological function.
Organizational Affiliation:
DISCAFF Department of Chemical, Food, Pharmaceutical and Pharmacological Sciences, University of Piemonte Orientale A Avogadro, 28100 Novara, Italy.