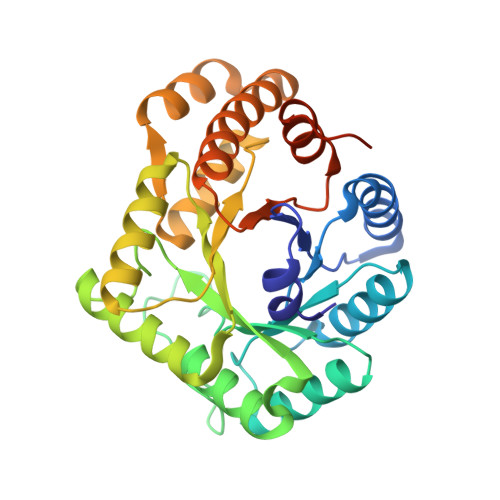
Properties and Crystal Structure of Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8.
Igari, S., Ohtaki, A., Yamanaka, Y., Sato, Y., Yohda, M., Odaka, M., Noguchi, K., Yamada, K.(2011) PLoS One 6: e23716-e23716
- PubMed: 21858212
- DOI: https://doi.org/10.1371/journal.pone.0023716
- Primary Citation of Related Structures:
3APT, 3APY - PubMed Abstract:
Methylenetetrahydrofolate reductase (MTHFR) is one of the enzymes involved in homocysteine metabolism. Despite considerable genetic and clinical attention, the reaction mechanism and regulation of this enzyme are not fully understood because of difficult production and poor stability. While recombinant enzymes from thermophilic organisms are often stable and easy to prepare, properties of thermostable MTHFRs have not yet been reported. MTHFR from Thermus thermophilus HB8, a homologue of Escherichia coli MetF, has been expressed in E. coli and purified. The purified MTHFR was chiefly obtained as a heterodimer of apo- and holo-subunits, that is, one flavin adenine dinucleotide (FAD) prosthetic group bound per dimer. The crystal structure of the holo-subunit was quite similar to the β(8)α(8) barrel of E. coli MTHFR, while that of the apo-subunit was a previously unobserved closed form. In addition, the intersubunit interface of the dimer in the crystals was different from any of the subunit interfaces of the tetramer of E. coli MTHFR. Free FAD could be incorporated into the apo-subunit of the purified Thermus enzyme after purification, forming a homodimer of holo-subunits. Comparison of the crystal structures of the heterodimer and the homodimer revealed different intersubunit interfaces, indicating a large conformational change upon FAD binding. Most of the biochemical properties of the heterodimer and the homodimer were the same, except that the homodimer showed ≈50% activity per FAD-bound subunit in folate-dependent reactions. The different intersubunit interfaces and rearrangement of subunits of Thermus MTHFR may be related to human enzyme properties, such as the allosteric regulation by S-adenosylmethionine and the enhanced instability of the Ala222Val mutant upon loss of FAD. Whereas E. coli MTHFR was the only structural model for human MTHFR to date, our findings suggest that Thermus MTHFR will be another useful model for this important enzyme.
Organizational Affiliation:
Department of Biotechnology and Life Science, Tokyo University of Agriculture and Technology, Koganei, Tokyo, Japan.