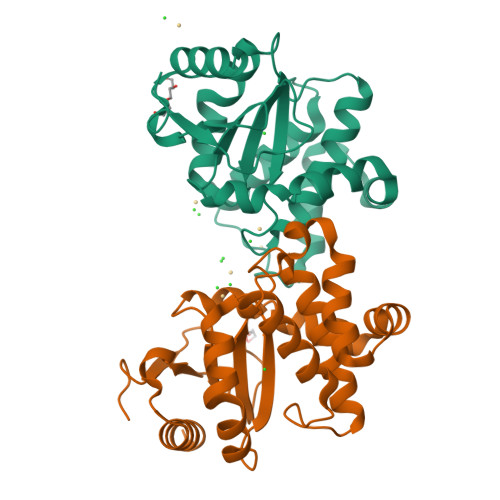
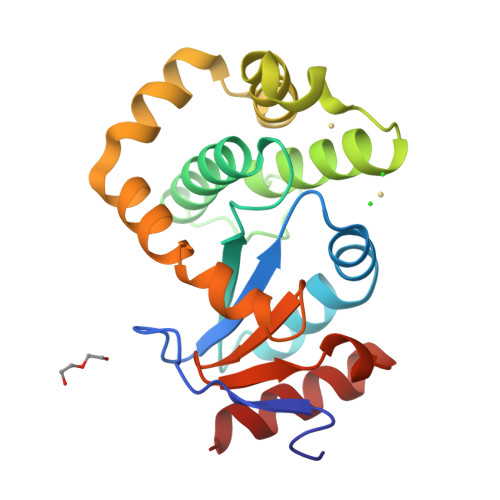
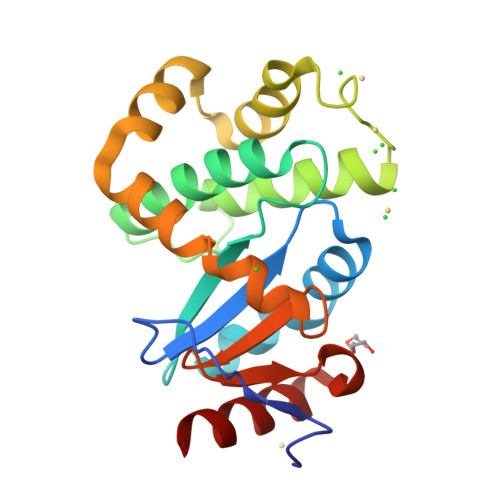DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli.
Grimshaw, J.P., Stirnimann, C.U., Brozzo, M.S., Malojcic, G., Grutter, M.G., Capitani, G., Glockshuber, R.(2008) J Mol Biology 380: 667-680
- PubMed: 18565543
- DOI: https://doi.org/10.1016/j.jmb.2008.05.031
- Primary Citation of Related Structures:
3C7M - PubMed Abstract:
Disulfide bond formation in the Escherichia coli periplasm requires the transfer of electrons from substrate proteins to DsbA, which is recycled as an oxidant by the membrane protein DsbB. The highly virulent, uropathogenic E. coli strain CFT073 contains a second, homologous pair of proteins, DsbL and DsbI, which are encoded in a tri-cistronic operon together with a periplasmic, uropathogen-specific arylsulfate sulfotransferase (ASST). We show that DsbL and DsbI form a functional redox pair, and that ASST is a substrate of DsbL/DsbI in vivo. DsbL is the most reactive oxidizing thioredoxin-like protein known to date. In contrast to DsbA, however, DsbL oxidizes reduced RNaseA with a much lower rate and prevents unspecific aggregation of reduced insulin. The 1.55 A resolution crystal structure of reduced DsbL provides insight into the reduced state of thioredoxin-like dithiol oxidases at high resolution, and reveals an unusual cluster of basic residues stabilizing the thiolate anion of the nucleophilic active-site cysteine. We propose that the DsbL/DsbI pair of uropathogenic E. coli was acquired as an additional, specific redox couple that guarantees biological activity of ASST.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, ETH Zürich, CH-8093 Zürich, Switzerland.