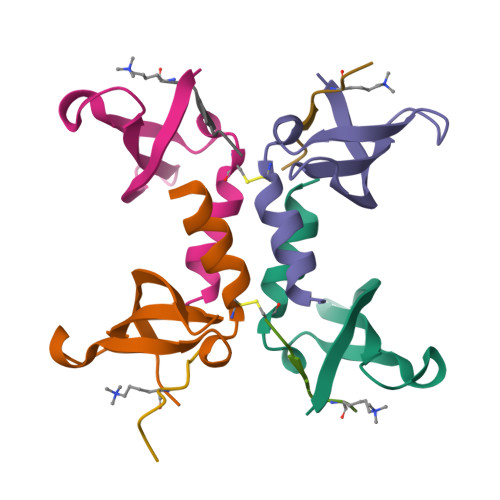
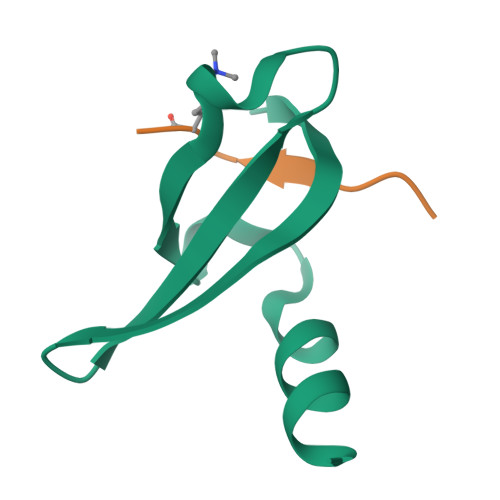
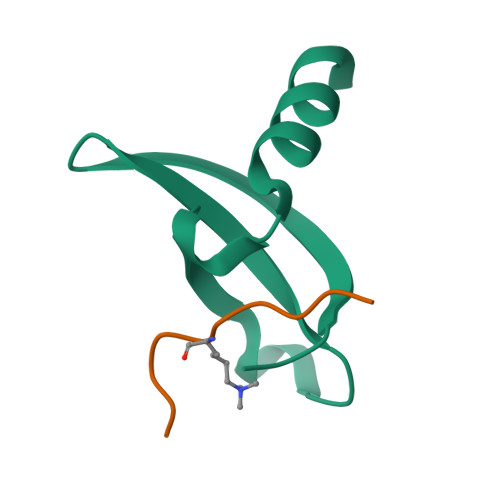
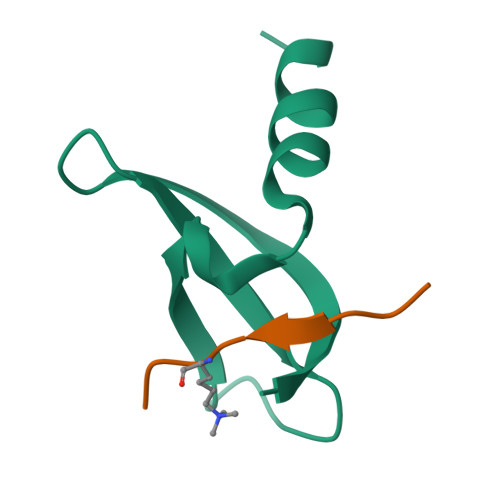
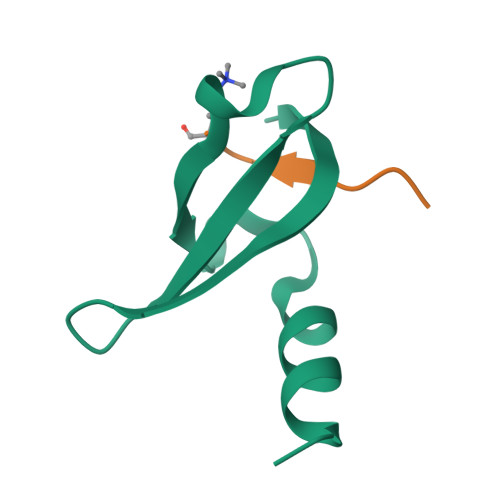
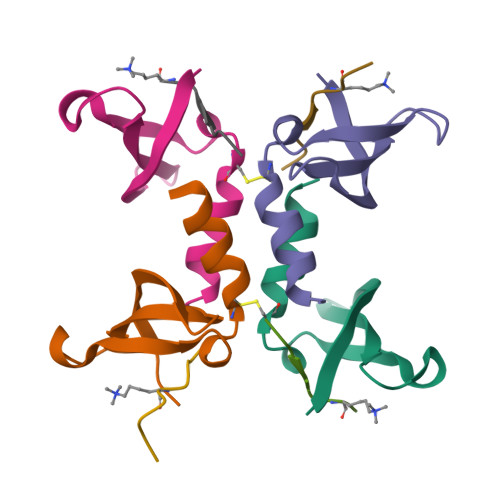
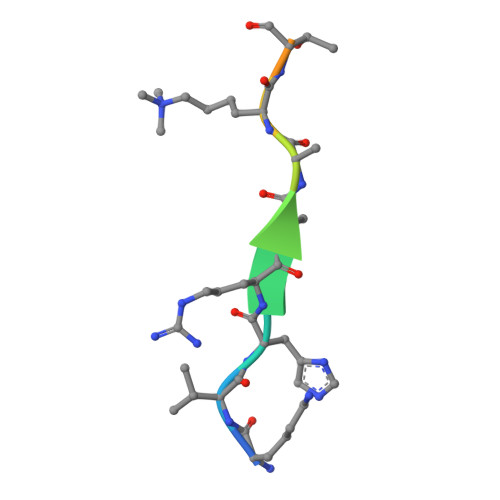
Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Ruan, J., Ouyang, H., Amaya, M.F., Ravichandran, M., Loppnau, P., Min, J., Zang, J.(2012) PLoS One 7: e35376-e35376
- PubMed: 22514736
- DOI: https://doi.org/10.1371/journal.pone.0035376
- Primary Citation of Related Structures:
3DM1, 3TZD - PubMed Abstract:
HP1 proteins are highly conserved heterochromatin proteins, which have been identified to be structural adapters assembling a variety of macromolecular complexes involved in regulation of gene expression, chromatin remodeling and heterochromatin formation. Much evidence shows that HP1 proteins interact with numerous proteins including methylated histones, histone methyltransferases and so on. Cbx3 is one of the paralogues of HP1 proteins, which has been reported to specifically recognize trimethylated histone H3K9 mark, and a consensus binding motif has been defined for the Cbx3 chromodomain. Here, we found that the Cbx3 chromodomain can bind to H1K26me2 and G9aK185me3 with comparable binding affinities compared to H3K9me3. We also determined the crystal structures of the human Cbx3 chromodomain in complex with dimethylated histone H1K26 and trimethylated G9aK185 peptides, respectively. The complex structures unveil that the Cbx3 chromodomain specifically bind methylated histone H1K26 and G9aK185 through a conserved mechanism. The Cbx3 chromodomain binds with comparable affinities to all of the methylated H3K9, H1K26 and G9aK185 peptides. It is suggested that Cbx3 may regulate gene expression via recognizing both histones and non-histone proteins.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences, Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, People's Republic of China.