Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae
Ngo, H.P.T., Kim, J.K., Kim, H.S., Jung, J.H., Ahn, Y.J., Kim, J.G., Lee, B.M., Kang, H.W., Kang, L.W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cystathionine gamma-lyase-like protein | 400 | Xanthomonas oryzae pv. oryzae | Mutation(s): 0 Gene Names: metB, XOO0778 | 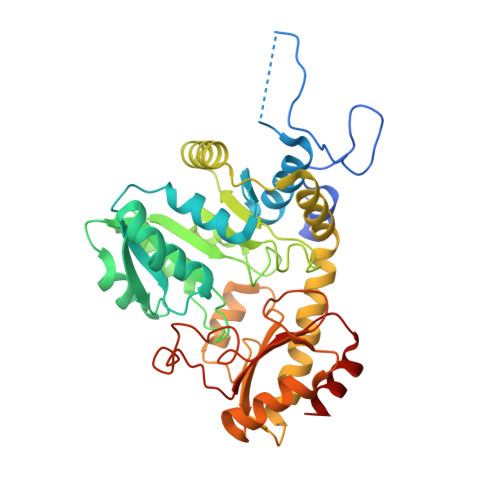 | |
UniProt | |||||
Find proteins for Q5H4T8 (Xanthomonas oryzae pv. oryzae (strain KACC10331 / KXO85)) Go to UniProtKB: Q5H4T8 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.351 | α = 90 |
| b = 78.351 | β = 90 |
| c = 300.989 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASES | phasing |