Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
Spadiut, O., Tan, T.C., Pisanelli, I., Haltrich, D., Divne, C.(2010) FEBS J 277: 2892-2909
- PubMed: 20528921
- DOI: https://doi.org/10.1111/j.1742-4658.2010.07705.x
- Primary Citation of Related Structures:
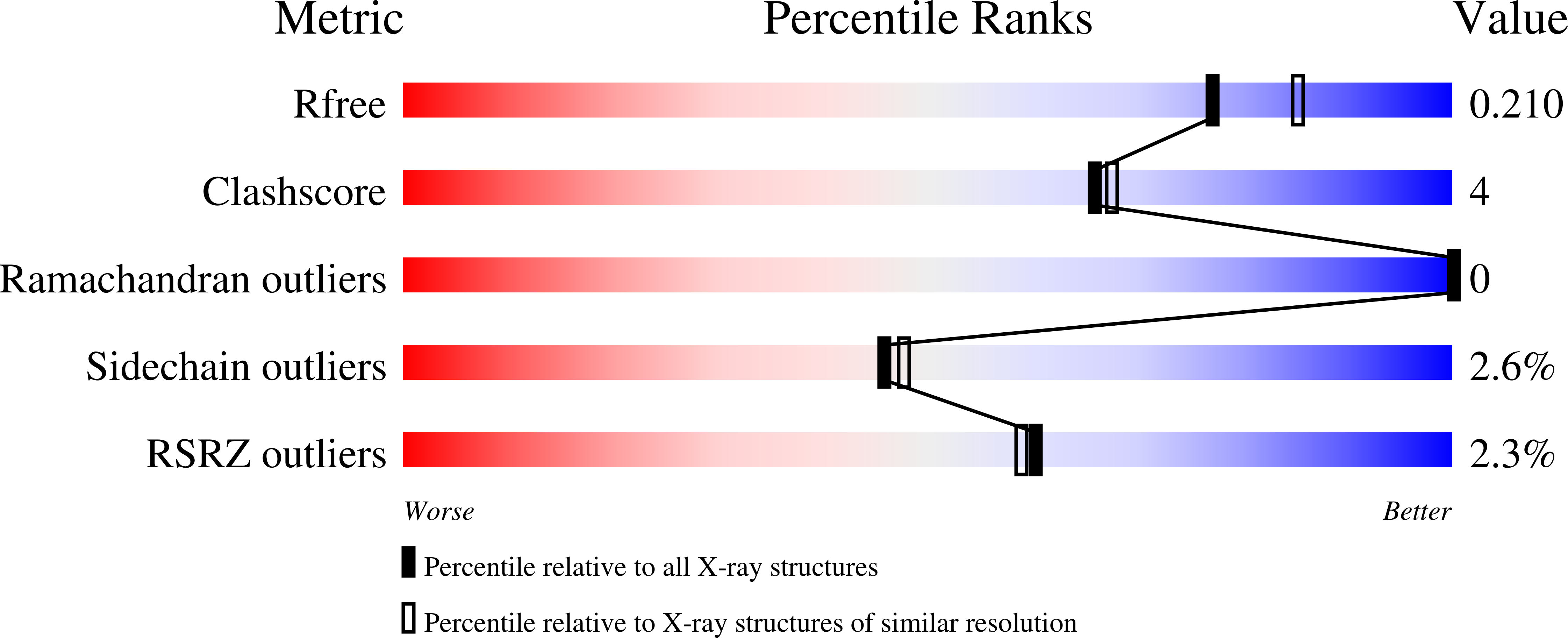
3K4J, 3K4K, 3K4L, 3K4M, 3K4N - PubMed Abstract:
Pyranose 2-oxidase from Trametes multicolor is a 270 kDa homotetrameric enzyme that participates in lignocellulose degradation by wood-rotting fungi and oxidizes a variety of aldopyranoses present in lignocellulose to 2-ketoaldoses. The active site in pyranose 2-oxidase is gated by a highly conserved, conformationally degenerate loop (residues 450-461), with a conformer ensemble that can accommodate efficient binding of both electron-donor substrate (sugar) and electron-acceptor substrate (oxygen or quinone compounds) relevant to the sequential reductive and oxidative half-reactions, respectively. To investigate the importance of individual residues in this loop, a systematic mutagenesis approach was used, including alanine-scanning, site-saturation and deletion mutagenesis, and selected variants were characterized by biochemical and crystal-structure analyses. We show that the gating segment ((454)FSY(456)) of this loop is particularly important for substrate specificity, discrimination of sugar substrates, turnover half-life and resistance to thermal unfolding, and that three conserved residues (Asp(452), Phe(454) and Tyr(456)) are essentially intolerant to substitution. We furthermore propose that the gating segment is of specific importance for the oxidative half-reaction of pyranose 2-oxidase when oxygen is the electron acceptor. Although the position and orientation of the slow substrate 2-deoxy-2-fluoro-glucose when bound in the active site of pyranose 2-oxidase variants is identical to that observed earlier, the substrate-recognition loop in F454N and Y456W displays a high degree of conformational disorder. The present study also lends support to the hypothesis that 1,4-benzoquinone is a physiologically relevant alternative electron acceptor in the oxidative half-reaction.
Organizational Affiliation:
KTH Biotechnology, Royal Institute of Technology, Stockholm, Sweden.