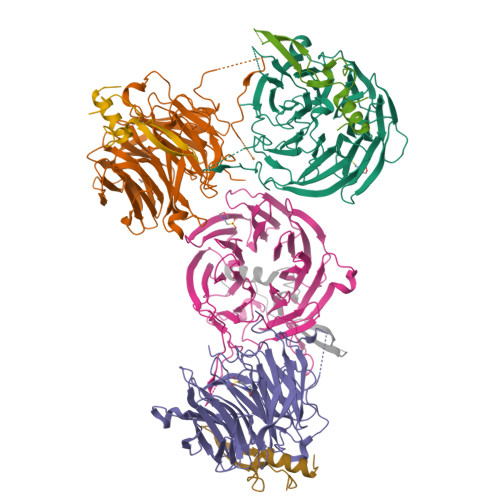
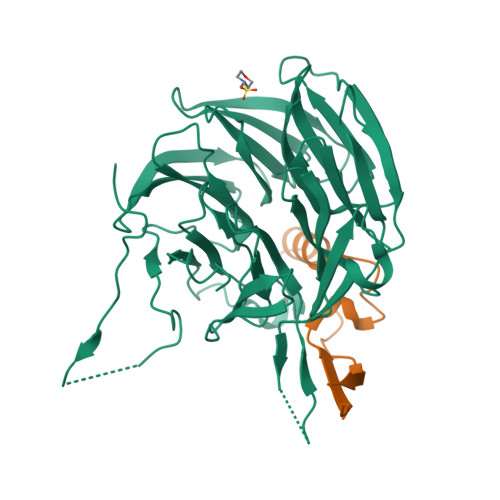
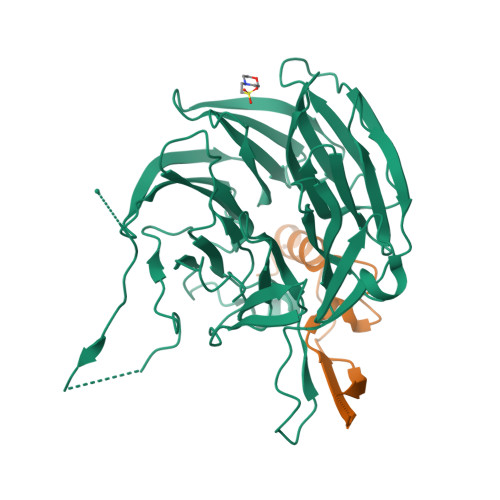
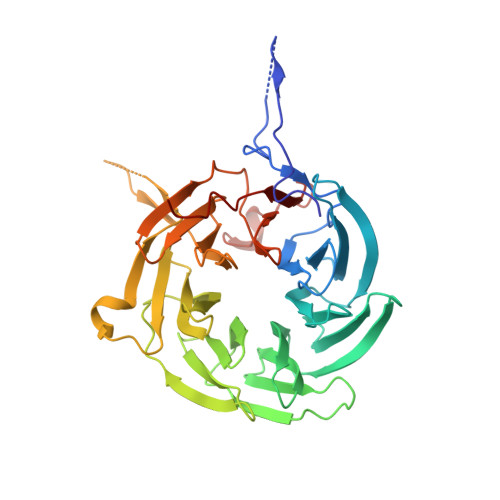
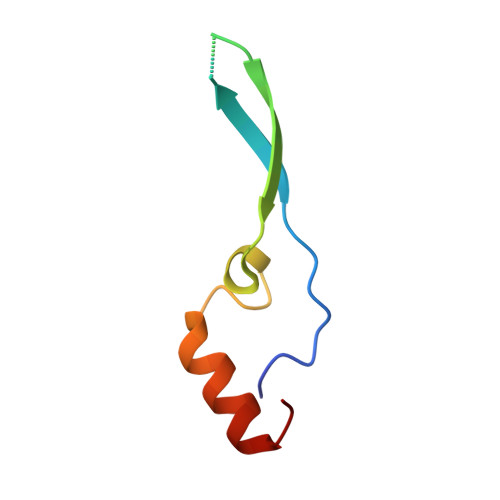
Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1.
Ren, Y., Seo, H.S., Blobel, G., Hoelz, A.(2010) Proc Natl Acad Sci U S A 107: 10406-10411
- PubMed: 20498086
- DOI: https://doi.org/10.1073/pnas.1005389107
- Primary Citation of Related Structures:
3MMY - PubMed Abstract:
The export of mRNAs is a multistep process, involving the packaging of mRNAs into messenger ribonucleoprotein particles (mRNPs), their transport through nuclear pore complexes, and mRNP remodeling events prior to translation. Ribonucleic acid export 1 (Rae1) and Nup98 are evolutionarily conserved mRNA export factors that are targeted by the vesicular stomatitis virus matrix protein to inhibit host cell nuclear export. Here, we present the crystal structure of human Rae1 in complex with the Gle2-binding sequence (GLEBS) of Nup98 at 1.65 A resolution. Rae1 forms a seven-bladed beta-propeller with several extensive surface loops. The Nup98 GLEBS motif forms an approximately 50-A-long hairpin that binds with its C-terminal arm to an essentially invariant hydrophobic surface that extends over the entire top face of the Rae1 beta-propeller. The C-terminal arm of the GLEBS hairpin is necessary and sufficient for Rae1 binding, and we identify a tandem glutamate element in this arm as critical for complex formation. The Rae1*Nup98(GLEBS) surface features an additional conserved patch with a positive electrostatic potential, and we demonstrate that the complex possesses single-stranded RNA-binding capability. Together, these data suggest that the Rae1*Nup98 complex directly binds to the mRNP at several stages of the mRNA export pathway.
Organizational Affiliation:
Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10065, USA.