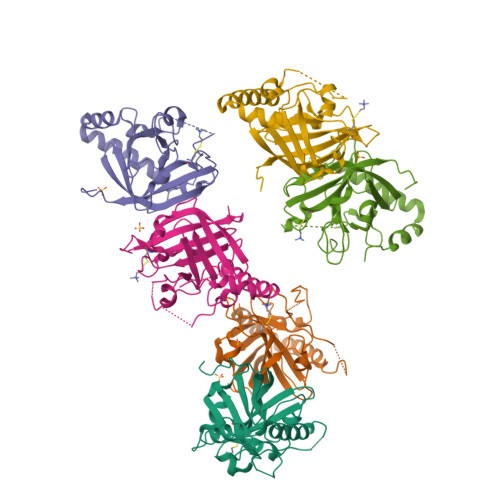
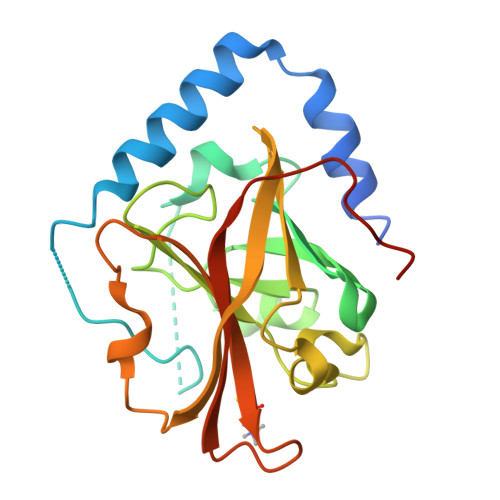
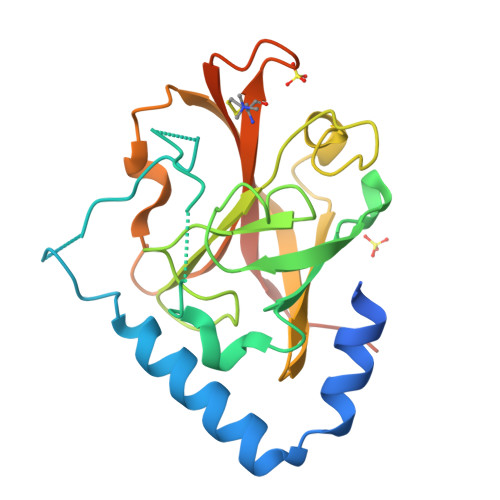
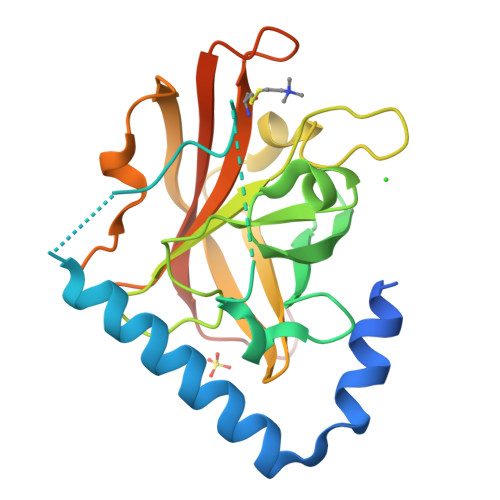
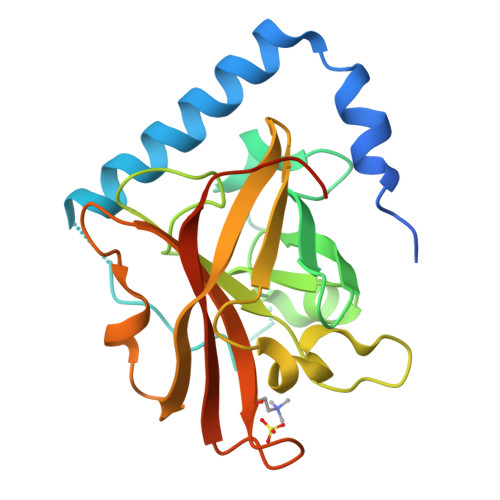
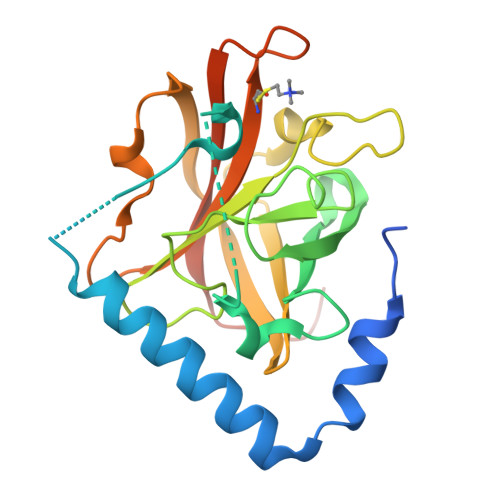
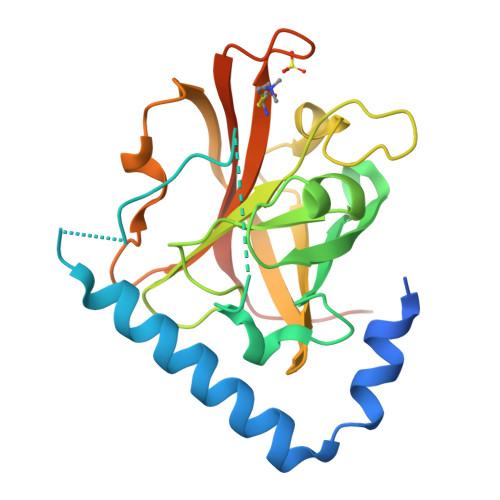
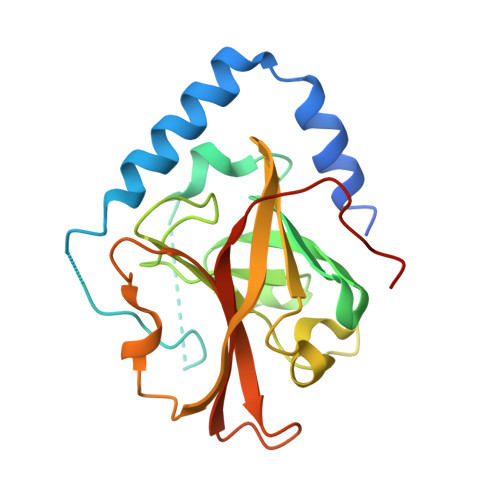
The Crystal Structure Analysis of Group B Streptococcus Sortase C1: A Model for the "Lid" Movement upon Substrate Binding.
Khare, B., Fu, Z.Q., Huang, I.H., Ton-That, H., Narayana, S.V.(2011) J Mol Biology 414: 563-577
- PubMed: 22033482
- DOI: https://doi.org/10.1016/j.jmb.2011.10.017
- Primary Citation of Related Structures:
3TB7, 3TBE - PubMed Abstract:
A unique feature of the class-C-type sortases, enzymes essential for Gram-positive pilus biogenesis, is the presence of a flexible "lid" anchored in the active site. However, the mechanistic details of the "lid" displacement, suggested to be a critical prelude for enzyme catalysis, are not yet known. This is partly due to the absence of enzyme-substrate and enzyme-inhibitor complex crystal structures. We have recently described the crystal structures of the Streptococcus agalactiae SAG2603 V/R sortase SrtC1 in two space groups (type II and type III) and that of its "lid" mutant and proposed a role of the "lid" as a protector of the active-site hydrophobic environment. Here, we report the crystal structures of SAG2603 V/R sortase C1 in a different space group (type I) and that of its complex with a small-molecule cysteine protease inhibitor. We observe that the catalytic Cys residue is covalently linked to the small-molecule inhibitor without lid displacement. However, the type I structure provides a view of the sortase SrtC1 lid displacement while having structural elements similar to a substrate sorting motif suitably positioned in the active site. We propose that these major conformational changes seen in the presence of a substrate mimic in the active site may represent universal features of class C sortase substrate recognition and enzyme activation.
Organizational Affiliation:
Center for Biophysical Sciences and Engineering, University of Alabama at Birmingham, Birmingham, AL 35294, USA.