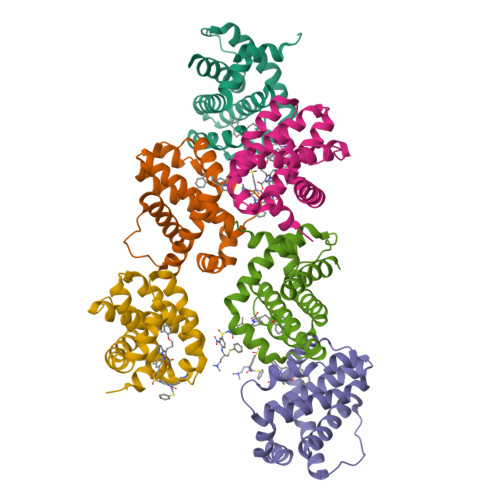
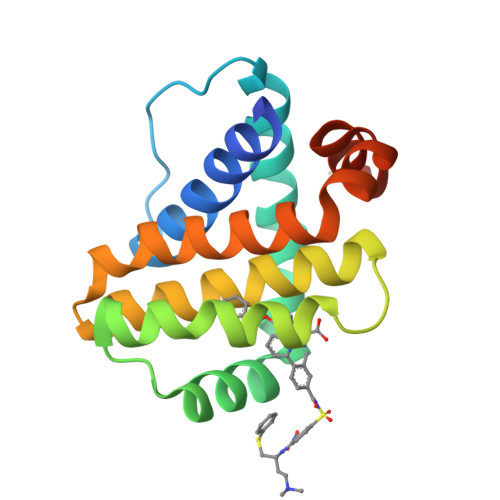
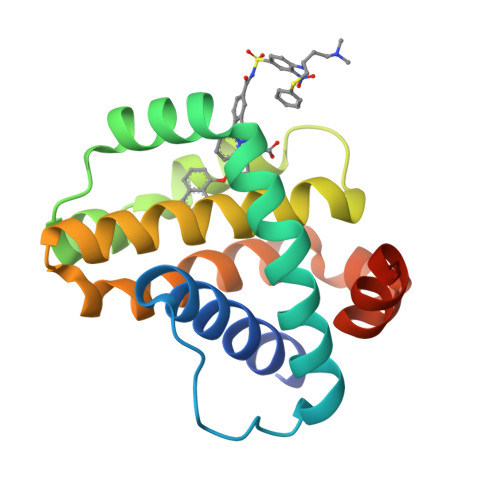
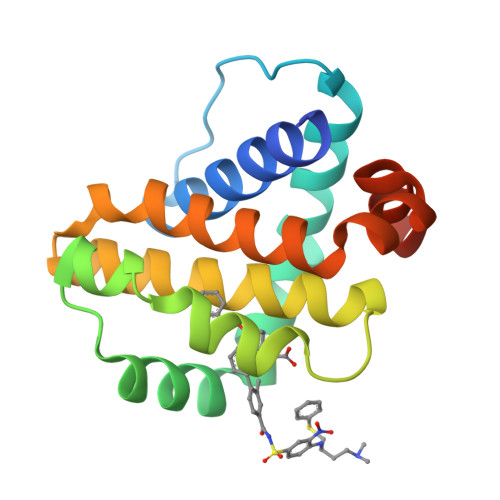
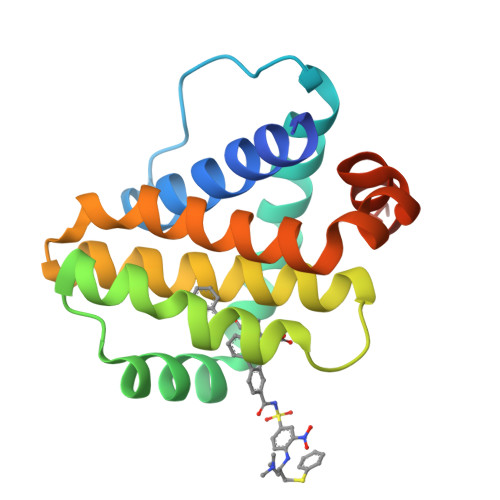
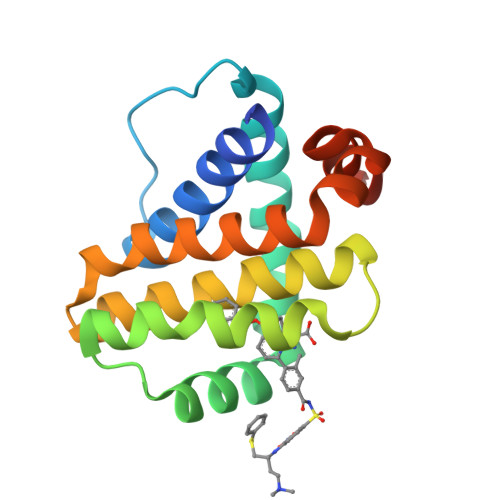
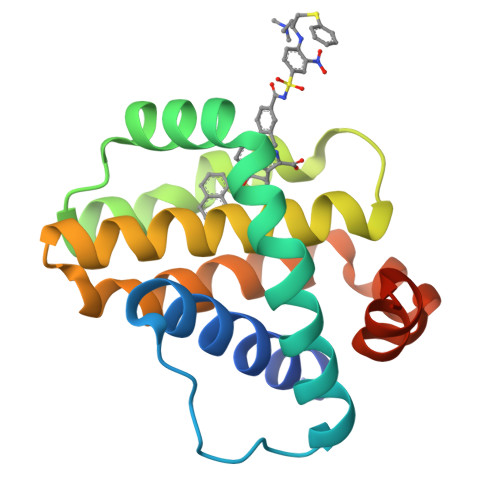
Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
Tanaka, Y., Aikawa, K., Nishida, G., Homma, M., Sogabe, S., Igaki, S., Hayano, Y., Sameshima, T., Miyahisa, I., Kawamoto, T., Tawada, M., Imai, Y., Inazuka, M., Cho, N., Imaeda, Y., Ishikawa, T.(2013) J Med Chem 56: 9635-9645
- PubMed: 24215352
- DOI: https://doi.org/10.1021/jm401170c
- Primary Citation of Related Structures:
3WIX, 3WIY, 3WIZ - PubMed Abstract:
Mcl-1 and Bcl-xL are crucial regulators of apoptosis, therefore dual inhibitors of both proteins could serve as promising new anticancer drugs. To design Mcl-1/Bcl-xL dual inhibitors, we performed structure-guided analyses of the corresponding selective Mcl-1 and Bcl-xL inhibitors. A cocrystal structure of a pyrazolo[1,5-a]pyridine derivative with Mcl-1 protein was successfully determined and revealed the protein-ligand binding mode. The key structure for Bcl-xL inhibition was further confirmed through the substructural analysis of ABT-263, a representative Bcl-xL/Bcl-2/Bcl-w inhibitor developed by Abbott Laboratories. On the basis of the structural data from this analysis, we designed hybrid compounds by tethering the Mcl-1 and Bcl-xL inhibitors together. The results of X-ray crystallographic analysis of hybrid compound 10 in complexes with both Mcl-1 and Bcl-xL demonstrated its binding mode with each protein. Following further optimization, compound 11 showed potent Mcl-1/Bcl-xL dual inhibitory activity (Mcl-1, IC50 = 0.088 μM; and Bcl-xL, IC50 = 0.0037 μM).
Organizational Affiliation:
Pharmaceutical Research Division, Takeda Pharmaceutical Company Limited , 26-1, Muraoka-Higashi 2-chome, Fujisawa, Kanagawa 251-8555, Japan.