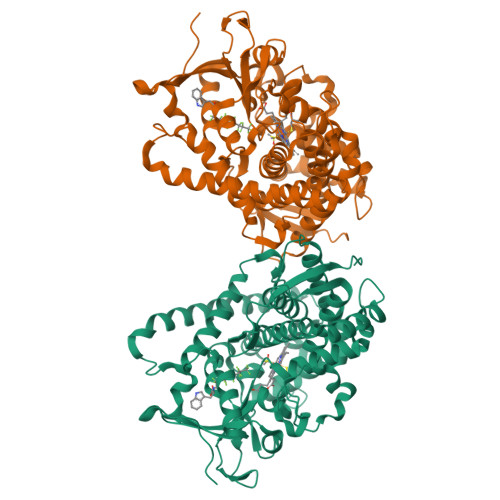
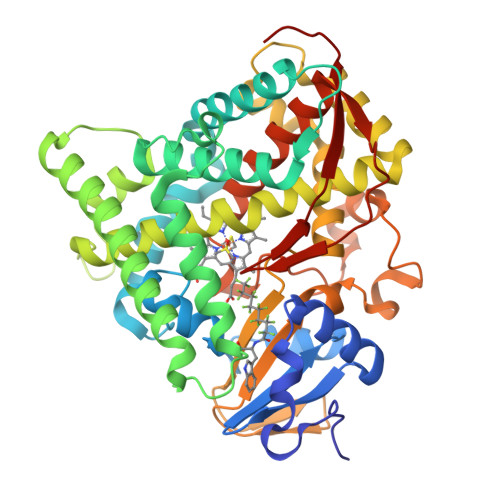
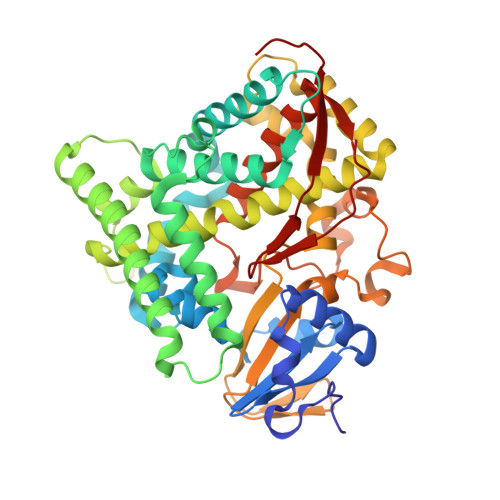
Activation of Wild-type Cytochrome P450BM3 by the Next Generation of Decoy Molecules: Enhanced Hydroxylation of Gaseous Alkanes and Crystallographic Evidence.
Cong, Z., Shoji, O., Kasai, C., Kawakami, N., Sugimoto, H., Shiro, Y., Watanabe, Y.(2015) ACS Catal 5: 150-156