The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Monteiro, D.C., Patel, V., Bartlett, C.P., Nozaki, S., Grant, T.D., Gowdy, J.A., Thompson, G.S., Kalverda, A.P., Snell, E.H., Niki, H., Pearson, A.R., Webb, M.E.(2015) Chem Biol 22: 492
- PubMed: 25910242
- DOI: https://doi.org/10.1016/j.chembiol.2015.03.017
- Primary Citation of Related Structures:
4CRZ, 4CS0 - PubMed Abstract:
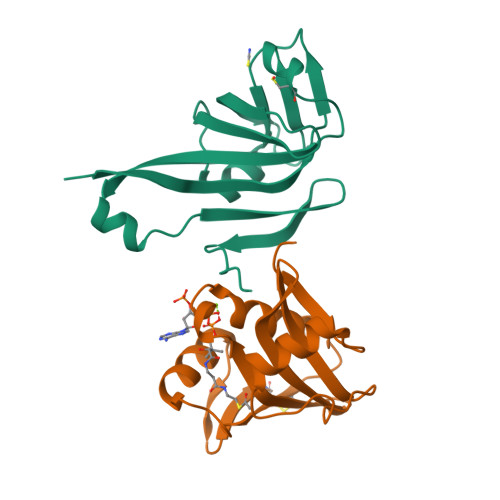
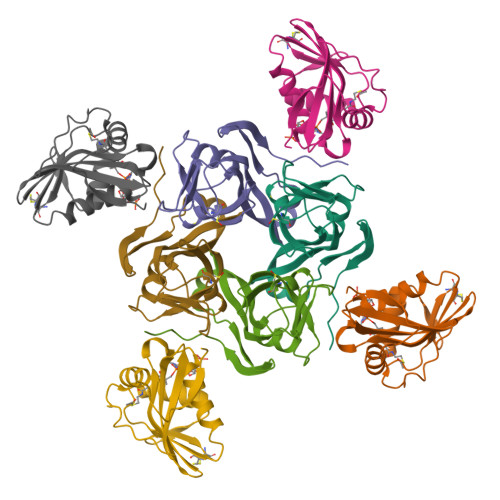
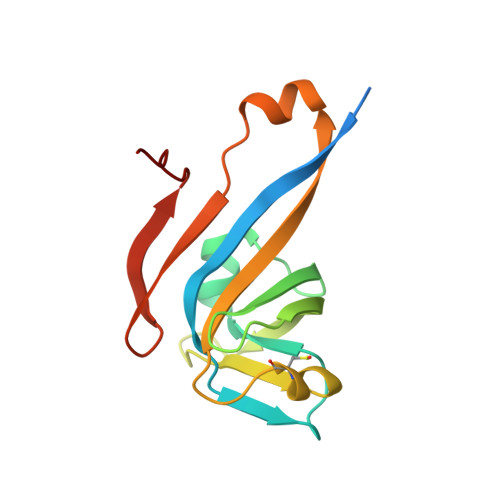
Coenzyme A (CoA) is an ubiquitous and essential cofactor, synthesized from the precursor pantothenate. Vitamin biosynthetic pathways are normally tightly regulated, including the pathway from pantothenate to CoA. However, no regulation of pantothenate biosynthesis has been identified. We have recently described an additional component in the pantothenate biosynthetic pathway, PanZ, which promotes the activation of the zymogen, PanD, to form aspartate α-decarboxylase (ADC) in a CoA-dependent manner. Here we report the structure of PanZ in complex with PanD, which reveals the structural basis for the CoA dependence of this interaction and activation. In addition, we show that PanZ acts as a CoA-dependent inhibitor of ADC catalysis. This inhibitory effect can effectively regulate the biosynthetic pathway to pantothenate, and thereby also regulate CoA biosynthesis. This represents a previously unobserved mode of metabolic regulation whereby a cofactor-utilizing protein negatively regulates the biosynthesis of the same cofactor.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, UK; School of Chemistry, University of Leeds, Leeds LS2 9JT, UK; Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, UK.