Structural basis of biological nitrile reduction.
Chikwana, V.M., Stec, B., Lee, B.W., de Crecy-Lagard, V., Iwata-Reuyl, D., Swairjo, M.A.(2012) J Biological Chem 287: 30560-30570
- PubMed: 22787148
- DOI: https://doi.org/10.1074/jbc.M112.388538
- Primary Citation of Related Structures:
4F8B, 4FGC - PubMed Abstract:
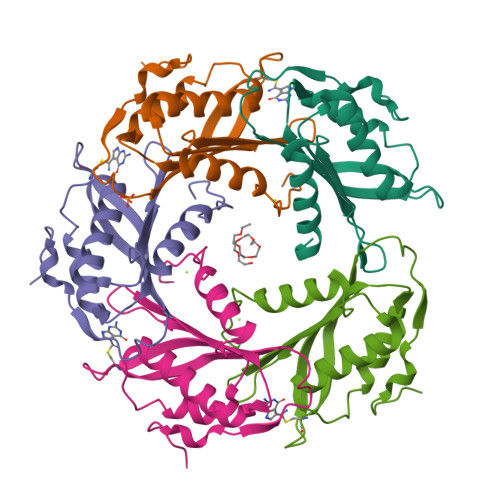
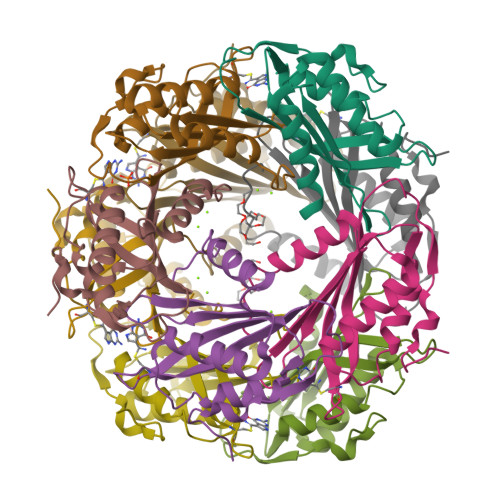
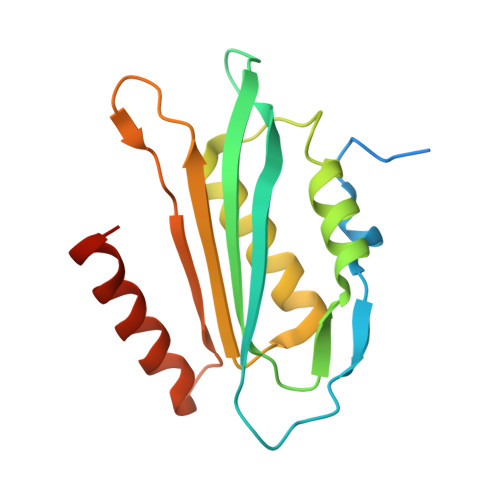
The enzyme QueF catalyzes the reduction of the nitrile group of 7-cyano-7-deazaguanine (preQ(0)) to 7-aminomethyl-7-deazaguanine (preQ(1)), the only nitrile reduction reaction known in biology. We describe here two crystal structures of Bacillus subtilis QueF, one of the wild-type enzyme in complex with the substrate preQ(0), trapped as a covalent thioimide, a putative intermediate in the reaction, and the second of the C55A mutant in complex with the substrate preQ(0) bound noncovalently. The QueF enzyme forms an asymmetric tunnel-fold homodecamer of two head-to-head facing pentameric subunits, harboring 10 active sites at the intersubunit interfaces. In both structures, a preQ(0) molecule is bound at eight sites, and in the wild-type enzyme, it forms a thioimide covalent linkage to the catalytic residue Cys-55. Both structural and transient kinetic data show that preQ(0) binding, not thioimide formation, induces a large conformational change in and closure of the active site. Based on these data, we propose a mechanism for the activation of the Cys-55 nucleophile and subsequent hydride transfer.
Organizational Affiliation:
Department of Chemistry, Portland State University, Portland, OR 97207, USA.