Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Liao, J.H., Ihara, K., Kuo, C.I., Huang, K.F., Wakatsuki, S., Wu, S.H., Chang, C.I.(2013) Acta Crystallogr D Biol Crystallogr 69: 1395-1402
- PubMed: 23897463
- DOI: https://doi.org/10.1107/S0907444913008214
- Primary Citation of Related Structures:
4FW9, 4FWD, 4FWG, 4FWH - PubMed Abstract:
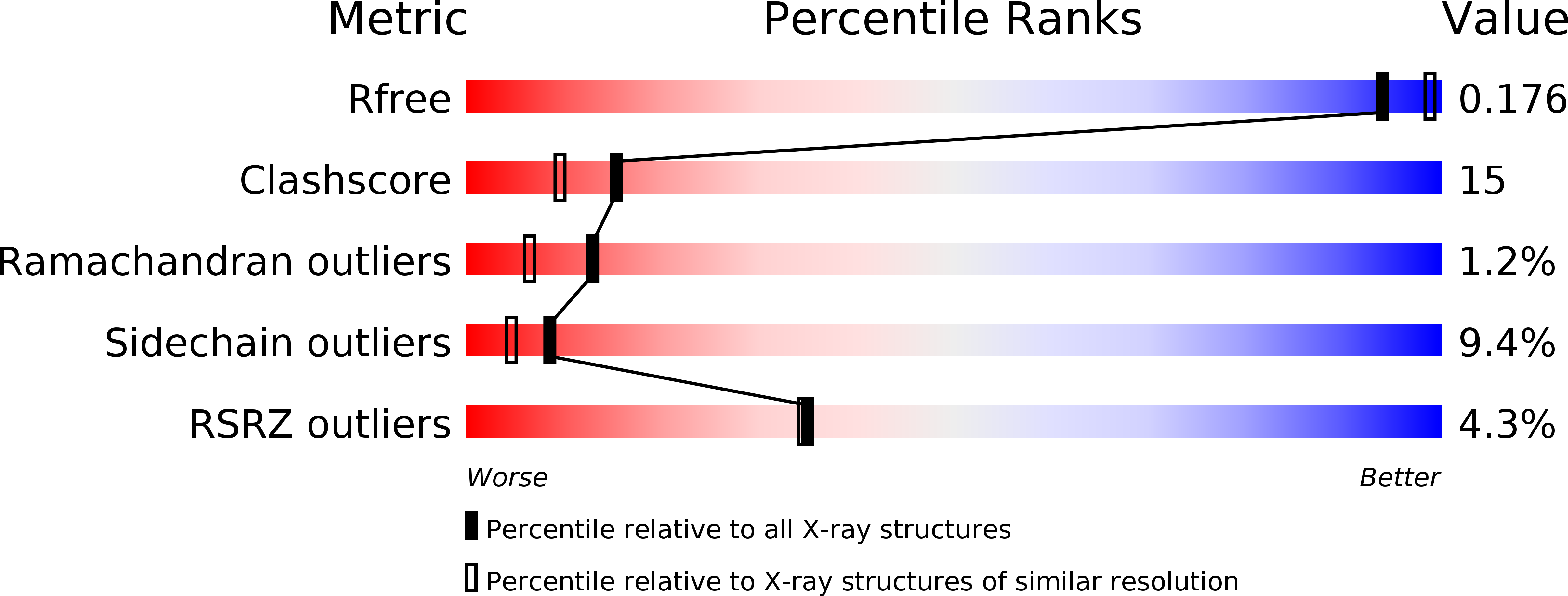
The Lon proteases are a unique family of chambered proteases with a built-in AAA+ (ATPases associated with diverse cellular activities) module. Here, crystal structures of a unique member of the Lon family with no intrinsic ATPase activity in the proteolytically active form are reported both alone and in complexes with three covalent inhibitors: two peptidomimetics and one derived from a natural product. This work reveals the unique architectural features of an ATP-independent Lon that selectively degrades unfolded protein substrates. Importantly, these results provide mechanistic insights into the recognition of inhibitors and polypeptide substrates within the conserved proteolytic chamber, which may aid the development of specific Lon-protease inhibitors.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan.