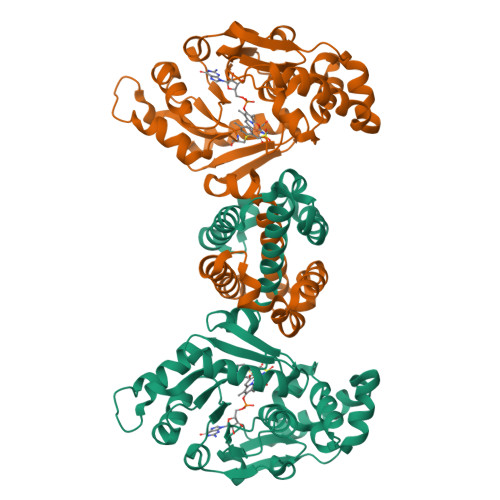
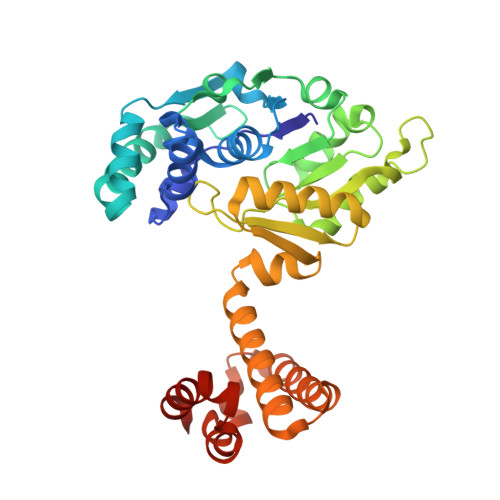
Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Tamura, H., Salomone-Stagni, M., Fujishiro, T., Warkentin, E., Meyer-Klaucke, W., Ermler, U., Shima, S.(2013) Angew Chem Int Ed Engl 52: 9656-9659
- PubMed: 23873755
- DOI: https://doi.org/10.1002/anie.201305089
- Primary Citation of Related Structures:
4JJF, 4JJG
Organizational Affiliation:
Max-Planck-Institut für terrestrische Mikrobiologie, Karl-von-Frisch-Strasse 10, 35043 Marburg, Germany.