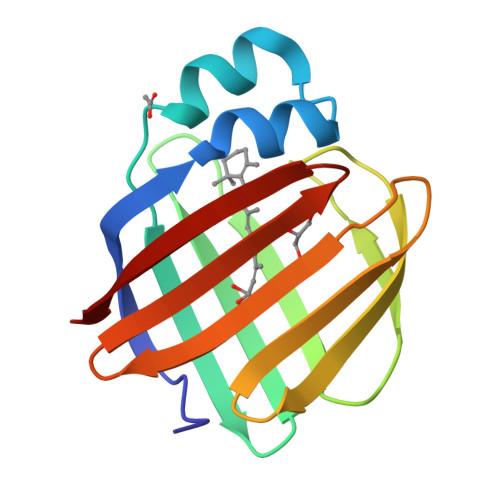
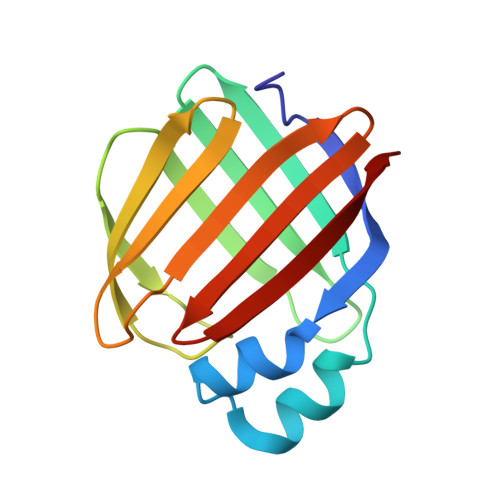
Structures of holo wild-type human cellular retinol-binding protein II (hCRBPII) bound to retinol and retinal.
Nossoni, Z., Assar, Z., Yapici, I., Nosrati, M., Wang, W., Berbasova, T., Vasileiou, C., Borhan, B., Geiger, J.(2014) Acta Crystallogr D Biol Crystallogr 70: 3226-3232
- PubMed: 25478840
- DOI: https://doi.org/10.1107/S1399004714023839
- Primary Citation of Related Structures:
4QYN, 4QYP, 4QZT, 4QZU - PubMed Abstract:
Cellular retinol-binding proteins (CRBPs) I and II, which are members of the intracellular lipid-binding protein (iLBP) family, are retinoid chaperones that are responsible for the intracellular transport and delivery of both retinol and retinal. Although structures of retinol-bound CRBPI and CRBPII are known, no structure of a retinal-bound CRBP has been reported. In addition, the retinol-bound human CRBPII (hCRBPII) structure shows partial occupancy of a noncanonical conformation of retinol in the binding pocket. Here, the structure of retinal-bound hCRBPII and the structure of retinol-bound hCRBPII with retinol fully occupying the binding pocket are reported. It is further shown that the retinoid derivative seen in both the zebrafish CRBP and the hCRBPII structures is likely to be the product of flux-dependent and wavelength-dependent X-ray damage during data collection. The structures of retinoid-bound CRBPs are compared and contrasted, and rationales for the differences in binding affinities for retinal and retinol are provided.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing, MI 48864, USA.