Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
Kuhnert, M., Blum, A., Steuber, H., Diederich, W.E.(2015) J Med Chem 58: 4845-4850
- PubMed: 26000468
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00346
- Primary Citation of Related Structures:
4YDF, 4YDG - PubMed Abstract:
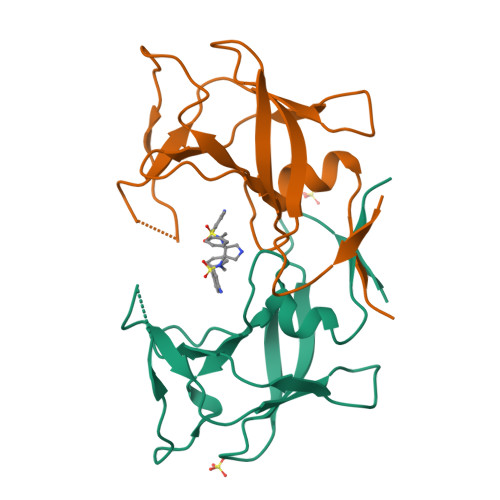
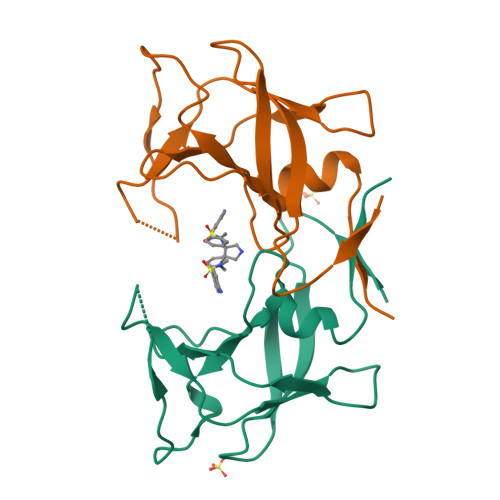
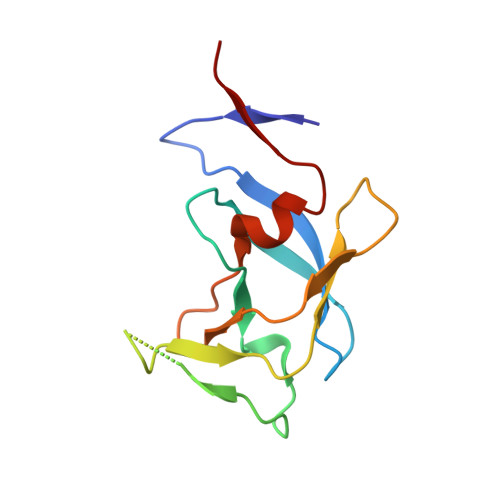
3,4-disubstituted pyrrolidines originally designed to inhibit the closely related HIV-1 protease were evaluated as privileged structures against HTLV-1 protease (HTLV-1 PR). The most potent inhibitor of this series exhibits two-digit nanomolar affinity and represents, to the best of our knowledge, the most potent nonpeptidic inhibitor of HTLV-1 PR described so far. The X-ray structures of two representatives bound to HTLV-1 PR were determined, and the structural basis of their affinity is discussed.
Organizational Affiliation:
†Institut für Pharmazeutische Chemie, Philipps-Universität Marburg, Hans-Meerwein-Straße 3, 35043 Marburg, Germany.