Structural Basis for the Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase by Sulfonamides.
Modakh, J.K., Liu, Y.C., Machuca, M.A., Supuran, C.T., Roujeinikova, A.(2015) PLoS One 10: e0127149-e0127149
- PubMed: 26010545
- DOI: https://doi.org/10.1371/journal.pone.0127149
- Primary Citation of Related Structures:
4YGF, 4YHA - PubMed Abstract:
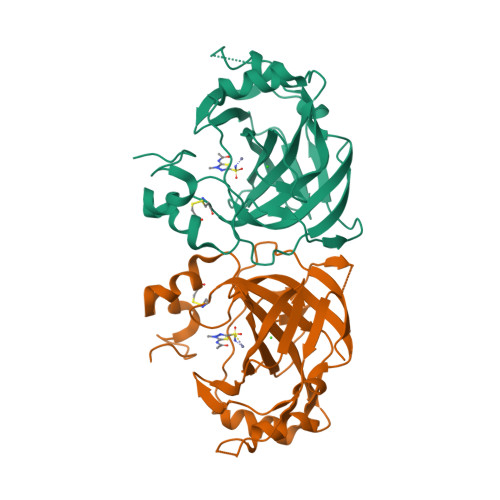
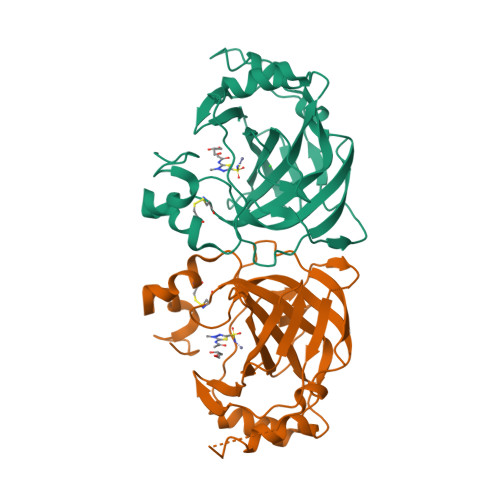
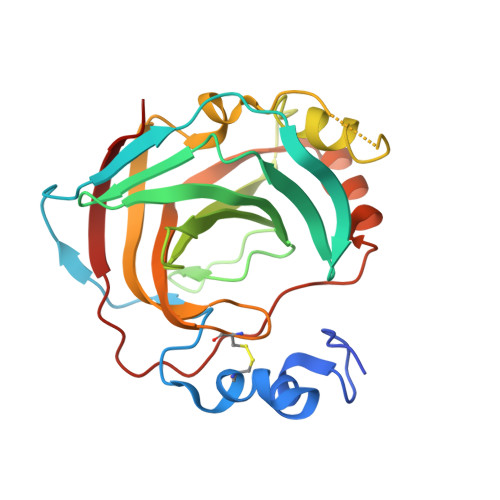
Periplasmic α-carbonic anhydrase of Helicobacter pylori (HpαCA), an oncogenic bacterium in the human stomach, is essential for its acclimation to low pH. It catalyses the conversion of carbon dioxide to bicarbonate using Zn(II) as the cofactor. In H. pylori, Neisseria spp., Brucella suis and Streptococcus pneumoniae this enzyme is the target for sulfonamide antibacterial agents. We present structural analysis correlated with inhibition data, on the complexes of HpαCA with two pharmacological inhibitors of human carbonic anhydrases, acetazolamide and methazolamide. This analysis reveals that two sulfonamide oxygen atoms of the inhibitors are positioned proximal to the putative location of the oxygens of the CO2 substrate in the Michaelis complex, whilst the zinc-coordinating sulfonamide nitrogen occupies the position of the catalytic water molecule. The structures are consistent with acetazolamide acting as site-directed, nanomolar inhibitors of the enzyme by mimicking its reaction transition state. Additionally, inhibitor binding provides insights into the channel for substrate entry and product exit. This analysis has implications for the structure-based design of inhibitors of bacterial carbonic anhydrases.
Organizational Affiliation:
Department of Microbiology, Faculty of Biomedical and Psychological Sciences, Monash University, Clayton, Victoria, Australia.