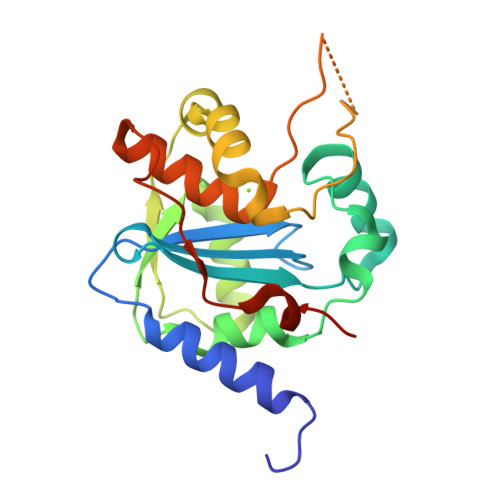
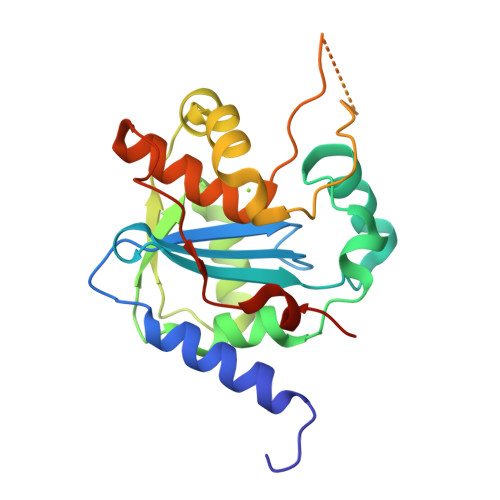
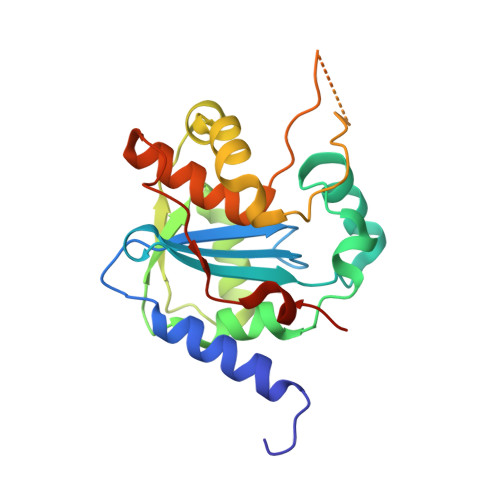
Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
Jiang, X., Huang, Q., Wang, W., Dong, H., Ly, H., Liang, Y., Dong, C.(2013) J Biological Chem 288: 16949-16959
- PubMed: 23615902
- DOI: https://doi.org/10.1074/jbc.M112.420521
- Primary Citation of Related Structures:
4G9Z, 4GV3, 4GV6, 4GV9, 4GVE - PubMed Abstract:
A hallmark of severe Lassa fever is the generalized immune suppression, the mechanism of which is poorly understood. Lassa virus (LASV) nucleoprotein (NP) is the only known 3'-5' exoribonuclease that can suppress type I interferon (IFN) production possibly by degrading immune-stimulatory RNAs. How this unique enzymatic activity of LASV NP recognizes and processes RNA substrates is unknown. We provide an atomic view of a catalytically active exoribonuclease domain of LASV NP (LASV NP-C) in the process of degrading a 5' triphosphate double-stranded (ds) RNA substrate, a typical pathogen-associated molecular pattern molecule, to induce type I IFN production. Additionally, we provide for the first time a high-resolution crystal structure of an active exoribonuclease domain of Tacaribe arenavirus (TCRV) NP. Coupled with the in vitro enzymatic and cell-based interferon suppression assays, these structural analyses strongly support a unified model of an exoribonuclease-dependent IFN suppression mechanism shared by all known arenaviruses. New knowledge learned from these studies should aid the development of therapeutics against pathogenic arenaviruses that can infect hundreds of thousands of individuals and kill thousands annually.
Organizational Affiliation:
Norwich Medical School, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, United Kingdom.