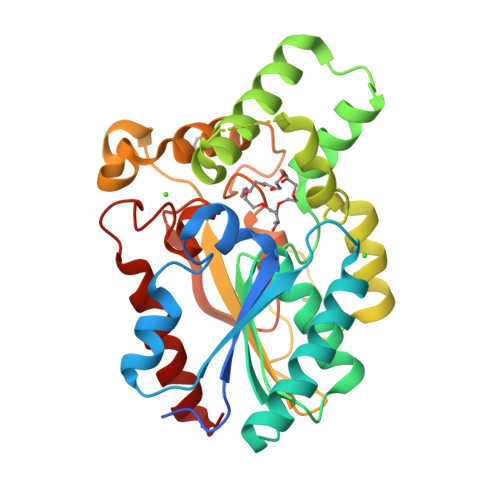
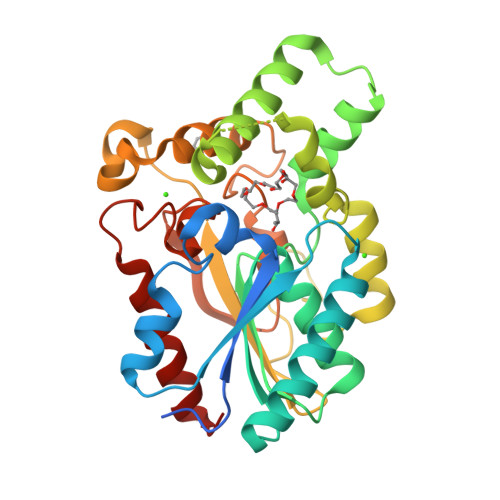
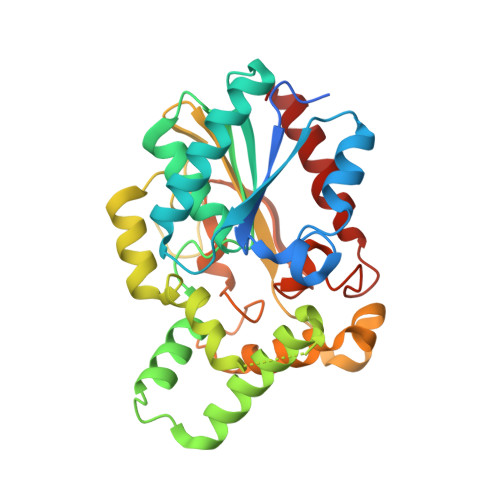
Crystal Structure of Proteus mirabilis Lipase, a Novel Lipase from the Proteus/Psychrophilic Subfamily of Lipase Family I.1.
Korman, T.P., Bowie, J.U.(2012) PLoS One 7: e52890-e52890
- PubMed: 23300806
- DOI: https://doi.org/10.1371/journal.pone.0052890
- Primary Citation of Related Structures:
4GW3, 4GXN - PubMed Abstract:
Bacterial lipases from family I.1 and I.2 catalyze the hydrolysis of triacylglycerol between 25-45°C and are used extensively as biocatalysts. The lipase from Proteus mirabilis belongs to the Proteus/psychrophilic subfamily of lipase family I.1 and is a promising catalyst for biodiesel production because it can tolerate high amounts of water in the reaction. Here we present the crystal structure of the Proteus mirabilis lipase, a member of the Proteus/psychrophilic subfamily of I.1lipases. The structure of the Proteus mirabilis lipase was solved in the absence and presence of a bound phosphonate inhibitor. Unexpectedly, both the apo and inhibitor bound forms of P. mirabilis lipase were found to be in a closed conformation. The structure reveals a unique oxyanion hole and a wide active site that is solvent accessible even in the closed conformation. A distinct mechanism for Ca²⁺ coordination may explain how these lipases can fold without specific chaperones.
Organizational Affiliation:
Department of Chemistry and Biochemisty, University of California Los Angeles, Los Angeles, California, United States of America.