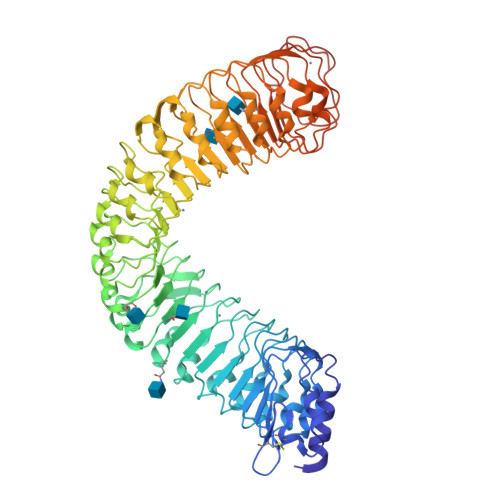
Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex.
Sun, Y., Li, L., Macho, A.P., Han, Z., Hu, Z., Zipfel, C., Zhou, J.M., Chai, J.(2013) Science 342: 624-628
- PubMed: 24114786
- DOI: https://doi.org/10.1126/science.1243825
- Primary Citation of Related Structures:
4MN8, 4MNA - PubMed Abstract:
Flagellin perception in Arabidopsis is through recognition of its highly conserved N-terminal epitope (flg22) by flagellin-sensitive 2 (FLS2). Flg22 binding induces FLS2 heteromerization with BRASSINOSTEROID INSENSITIVE 1-associated kinase 1 (BAK1) and their reciprocal activation followed by plant immunity. Here, we report the crystal structure of FLS2 and BAK1 ectodomains complexed with flg22 at 3.06 angstroms. A conserved and a nonconserved site from the inner surface of the FLS2 solenoid recognize the C- and N-terminal segment of flg22, respectively, without oligomerization or conformational changes in the FLS2 ectodomain. Besides directly interacting with FLS2, BAK1 acts as a co-receptor by recognizing the C terminus of the FLS2-bound flg22. Our data reveal the molecular mechanisms underlying FLS2-BAK1 complex recognition of flg22 and provide insight into the immune receptor complex activation.
Organizational Affiliation:
School of Life Sciences, Tsinghua University, Beijing 100084, China, and Tsinghua-Peking Center for Life Sciences, Beijing 100084, China.