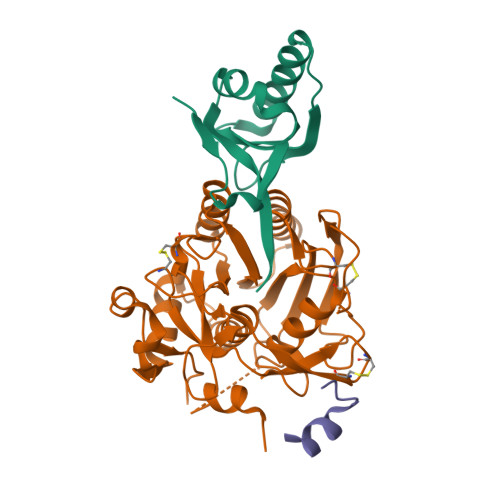
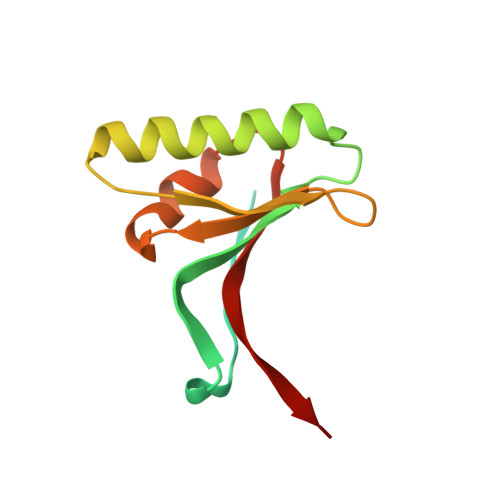
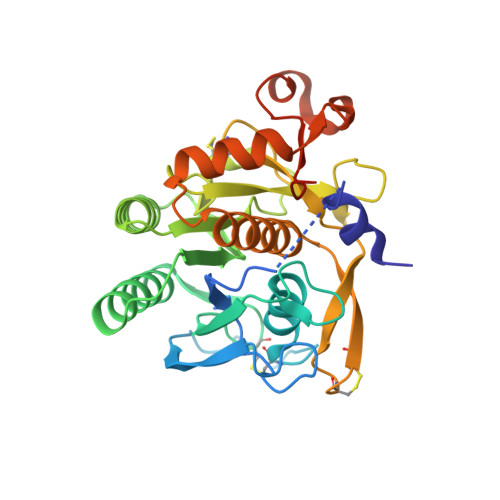
Identification of a Small Peptide That Inhibits PCSK9 Protein Binding to the Low Density Lipoprotein Receptor.
Zhang, Y., Eigenbrot, C., Zhou, L., Shia, S., Li, W., Quan, C., Tom, J., Moran, P., Di Lello, P., Skelton, N.J., Kong-Beltran, M., Peterson, A., Kirchhofer, D.(2014) J Biological Chem 289: 942-955
- PubMed: 24225950
- DOI: https://doi.org/10.1074/jbc.M113.514067
- Primary Citation of Related Structures:
4NMX - PubMed Abstract:
PCSK9 (proprotein convertase subtilisin/kexin type 9) is a negative regulator of the hepatic LDL receptor, and clinical studies with PCSK9-inhibiting antibodies have demonstrated strong LDL-c-lowering effects. Here we screened phage-displayed peptide libraries and identified the 13-amino acid linear peptide Pep2-8 as the smallest PCSK9 inhibitor with a clearly defined mechanism of inhibition that has been described. Pep2-8 bound to PCSK9 with a KD of 0.7 μm but did not bind to other proprotein convertases. It fully restored LDL receptor surface levels and LDL particle uptake in PCSK9-treated HepG2 cells. The crystal structure of Pep2-8 bound to C-terminally truncated PCSK9 at 1.85 Å resolution showed that the peptide adopted a strand-turn-helix conformation, which is remarkably similar to its solution structure determined by NMR. Consistent with the functional binding site identified by an Ala scan of PCSK9, the structural Pep2-8 contact region of about 400 Å(2) largely overlapped with that contacted by the EGF(A) domain of the LDL receptor, suggesting a competitive inhibition mechanism. Consistent with this, Pep2-8 inhibited LDL receptor and EGF(A) domain binding to PCSK9 with IC50 values of 0.8 and 0.4 μm, respectively. Remarkably, Pep2-8 mimicked secondary structural elements of the EGF(A) domain that interact with PCSK9, notably the β-strand and a discontinuous short α-helix, and it engaged in the same β-sheet hydrogen bonds as EGF(A) does. Although Pep2-8 itself may not be amenable to therapeutic applications, this study demonstrates the feasibility of developing peptidic inhibitors to functionally relevant sites on PCSK9.
Organizational Affiliation:
From the Departments of Early Discovery Biochemistry.