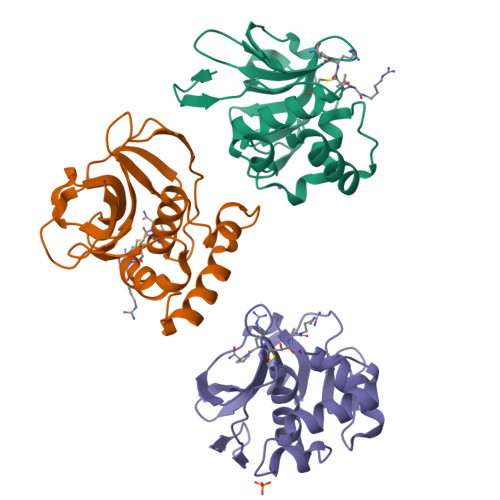
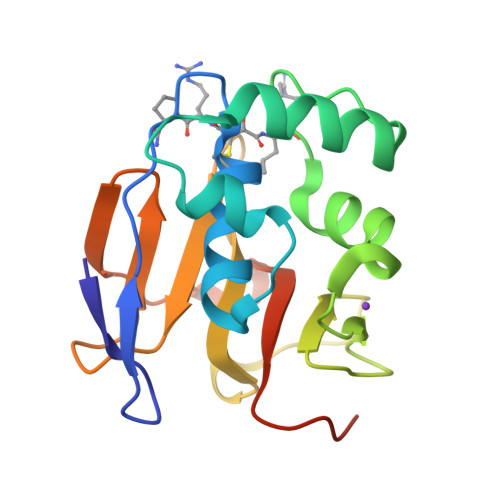
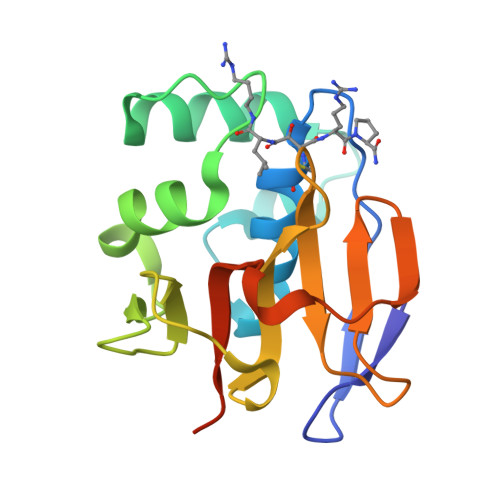
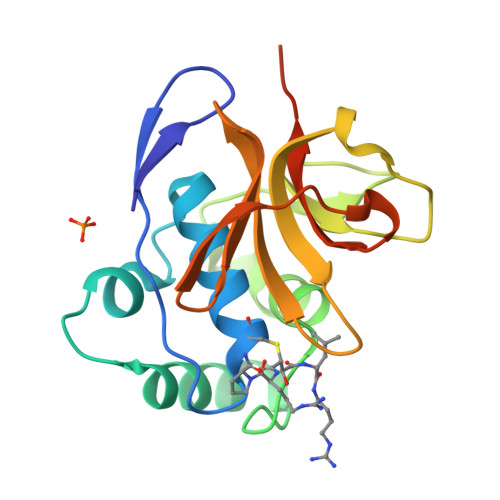
Foot-and-mouth disease virus leader proteinase: Structural insights into the mechanism of intermolecular cleavage.
Steinberger, J., Grishkovskaya, I., Cencic, R., Juliano, L., Juliano, M.A., Skern, T.(2014) Virology 468-470C: 397-408
- PubMed: 25240326
- DOI: https://doi.org/10.1016/j.virol.2014.08.023
- Primary Citation of Related Structures:
4QBB - PubMed Abstract:
Translation of foot-and-mouth disease virus RNA initiates at one of two start codons leading to the synthesis of two forms of leader proteinase L(pro) (Lab(pro) and Lb(pro)). These forms free themselves from the viral polyprotein by intra- and intermolecular self-processing and subsequently cleave the cellular eukaryotic initiation factor (eIF) 4 G. During infection, Lb(pro) removes six residues from its own C-terminus, generating sLb(pro). We present the structure of sLb(pro) bound to the inhibitor E64-R-P-NH2, illustrating how sLb(pro) can cleave between Lys/Gly and Gly/Arg pairs. In intermolecular cleavage on polyprotein substrates, Lb(pro) was unaffected by P1 or P1' substitutions and processed a substrate containing nine eIF4GI cleavage site residues whereas sLb(pro) failed to cleave the eIF4GI containing substrate and cleaved appreciably more slowly on mutated substrates. Introduction of 70 eIF4GI residues bearing the Lb(pro) binding site restored cleavage. These data imply that Lb(pro) and sLb(pro) may have different functions in infected cells.
Organizational Affiliation:
Max F. Perutz Laboratories, Medical University of Vienna, Dr. Bohr-Gasse 9/3, A-1030 Vienna, Austria.