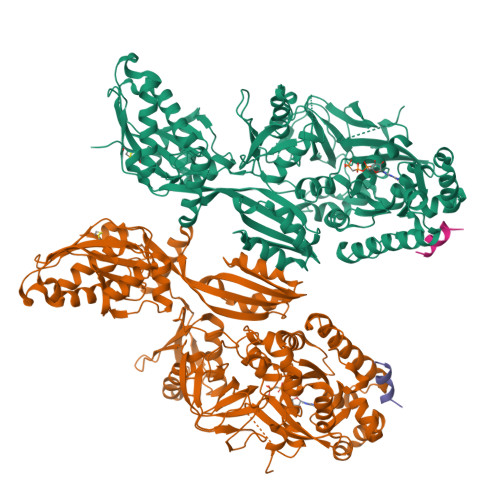
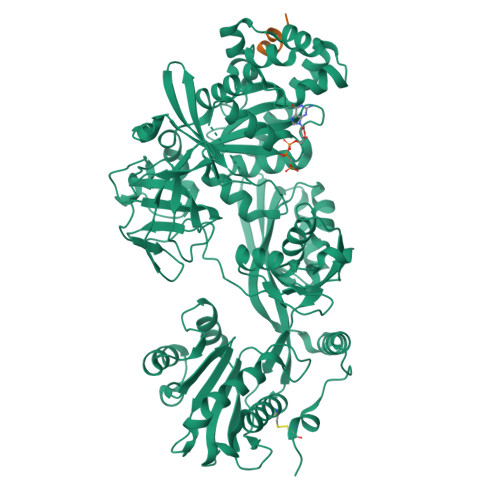
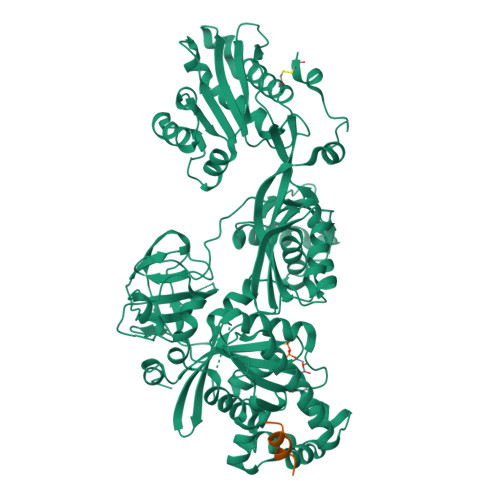
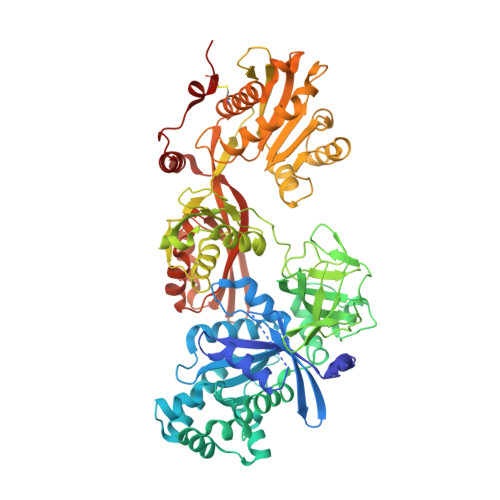
The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Tanzawa, T., Kato, K., Girodat, D., Ose, T., Kumakura, Y., Wieden, H.J., Uchiumi, T., Tanaka, I., Yao, M.(2018) Nucleic Acids Res 46: 3232-3244
- PubMed: 29471537
- DOI: https://doi.org/10.1093/nar/gky115
- Primary Citation of Related Structures:
5H7J, 5H7K, 5H7L - PubMed Abstract:
Archaea and eukaryotes have ribosomal P stalks composed of anchor protein P0 and aP1 homodimers (archaea) or P1•P2 heterodimers (eukaryotes). These P stalks recruit translational GTPases to the GTPase-associated center in ribosomes to provide energy during translation. The C-terminus of the P stalk is known to selectively recognize GTPases. Here we investigated the interaction between the P stalk and elongation factor 2 by determining the structures of Pyrococcus horikoshii EF-2 (PhoEF-2) in the Apo-form, GDP-form, GMPPCP-form (GTP-form), and GMPPCP-form bound with 11 C-terminal residues of P1 (P1C11). Helical structured P1C11 binds to a hydrophobic groove between domain G and subdomain G' of PhoEF-2, where is completely different from that of aEF-1α in terms of both position and sequence, implying that such interaction characteristic may be requested by how GTPases perform their functions on the ribosome. Combining PhoEF-2 P1-binding assays with a structural comparison of current PhoEF-2 structures and molecular dynamics model of a P1C11-bound GDP form, the conformational changes of the P1C11-binding groove in each form suggest that in response to the translation process, the groove has three states: closed, open, and release for recruiting and releasing GTPases.
Organizational Affiliation:
Graduate School of Life Science, Hokkaido University, Sapporo, Hokkaido 060-0810, Japan.