TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma.
Nakayama, K., Szewczyk, M.M., Dela Sena, C., Wu, H., Dong, A., Zeng, H., Li, F., de Freitas, R.F., Eram, M.S., Schapira, M., Baba, Y., Kunitomo, M., Cary, D.R., Tawada, M., Ohashi, A., Imaeda, Y., Saikatendu, K.S., Grimshaw, C.E., Vedadi, M., Arrowsmith, C.H., Barsyte-Lovejoy, D., Kiba, A., Tomita, D., Brown, P.J.(2018) Oncotarget 9: 18480-18493
- PubMed: 29719619
- DOI: https://doi.org/10.18632/oncotarget.24883
- Primary Citation of Related Structures:
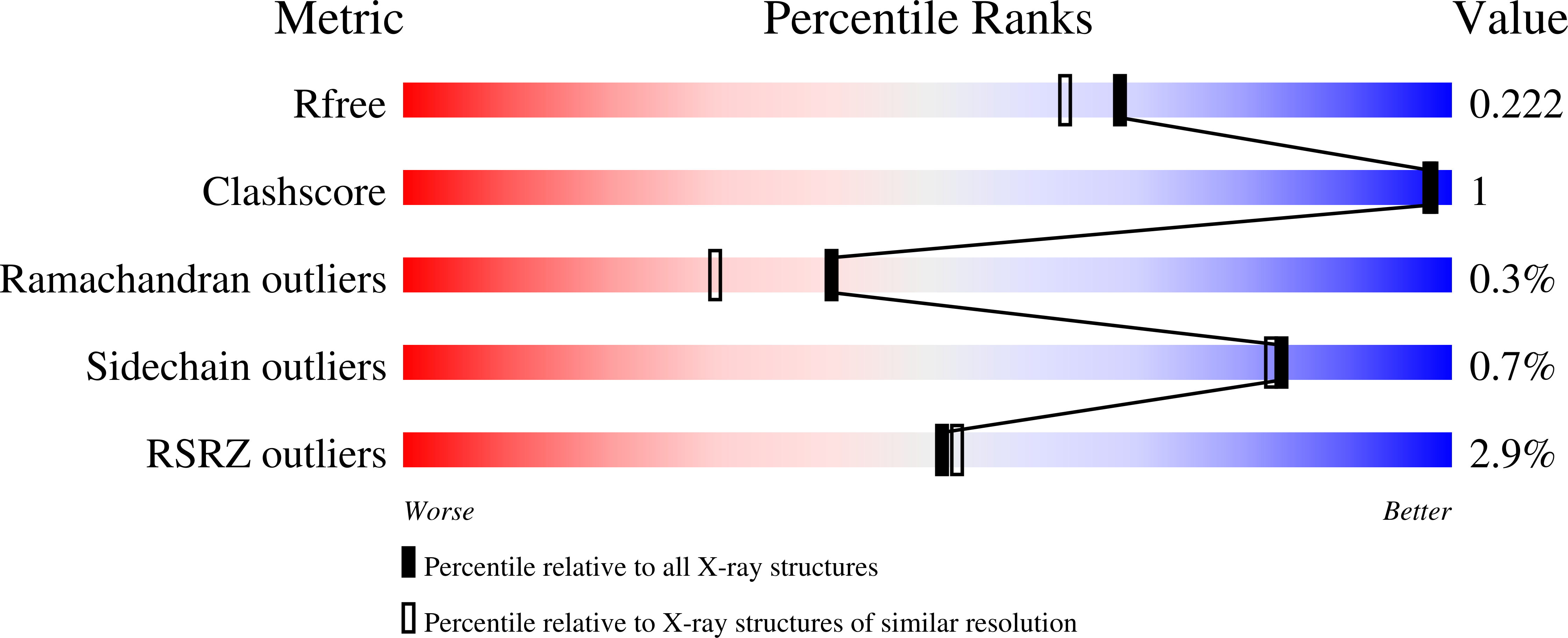
5U4X - PubMed Abstract:
Protein arginine methyltransferase (PRMT) 4 (also known as coactivator-associated arginine methyltransferase 1; CARM1) is involved in a variety of biological processes and is considered as a candidate oncogene owing to its overexpression in several types of cancer. Selective PRMT4 inhibitors are useful tools for clarifying the molecular events regulated by PRMT4 and for validating PRMT4 as a therapeutic target. Here, we report the discovery of TP-064, a potent, selective, and cell-active chemical probe of human PRMT4 and its co-crystal structure with PRMT4. TP-064 inhibited the methyltransferase activity of PRMT4 with high potency (half-maximal inhibitory concentration, IC 50 < 10 nM) and selectivity over other PRMT family proteins, and reduced arginine dimethylation of the PRMT4 substrates BRG1-associated factor 155 (BAF155; IC 50 = 340 ± 30 nM) and Mediator complex subunit 12 (MED12; IC 50 = 43 ± 10 nM). TP-064 treatment inhibited the proliferation of a subset of multiple myeloma cell lines, with affected cells arrested in G1 phase of the cell cycle. TP-064 and its negative control (TP-064N) will be valuable tools to further investigate the biology of PRMT4 and the therapeutic potential of PRMT4 inhibition.
Organizational Affiliation:
Oncology Drug Discovery Unit, Pharmaceutical Research Division, Takeda Pharmaceutical Company Limited, Fujisawa, Kanagawa 251-8555, Japan.