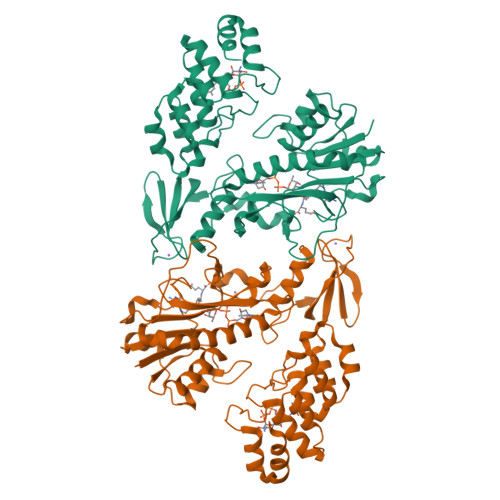
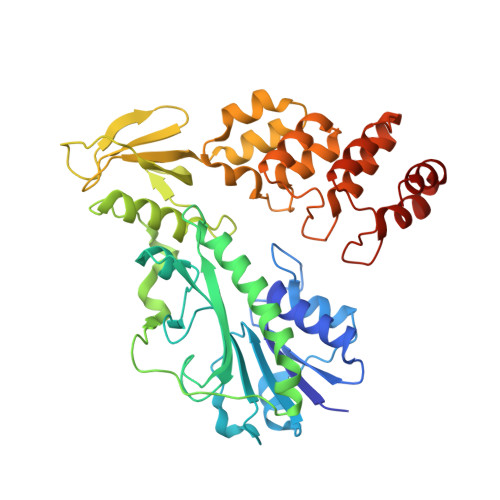
Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
Woodford, C.R., Thoden, J.B., Holden, H.M.(2017) J Struct Biol 200: 267-278
- PubMed: 28263875
- DOI: https://doi.org/10.1016/j.jsb.2017.03.002
- Primary Citation of Related Structures:
5UIJ, 5UIK, 5UIL, 5UIM, 5UIN - PubMed Abstract:
N-formylated sugars are found on the lipopolysaccharides of various pathogenic Gram negative bacteria including Campylobacter jejuni 81116, Francisella tularensis, Providencia alcalifaciens O30, and Providencia alcalifaciens O40. The last step in the biosynthetic pathways for these unusual sugars is catalyzed by N-formyltransferases that utilize N 10 -formyltetrahydrofolate as the carbon source. The substrates are dTDP-linked amino sugars with the functional groups installed at either the C-3' or C-4' positions of the pyranosyl rings. Here we describe a structural and enzymological investigation of the putative N-formyltransferase, FdtF, from Salmonella enterica O60. In keeping with its proposed role in the organism, the kinetic data reveal that the enzyme is more active with dTDP-3-amino-3,6-dideoxy-d-galactose than with dTDP-3-amino-3,6-dideoxy-d-glucose. The structural data demonstrate that the enzyme contains, in addition to the canonical N-formyltransferase fold, an ankyrin repeat moiety that houses a second dTDP-sugar binding pocket. This is only the second time an ankyrin repeat has been shown to be involved in small molecule binding. The research described herein represents the first structural analysis of a sugar N-formyltransferase that specifically functions on dTDP-3-amino-3,6-dideoxy-d-galactose in vivo and thus adds to our understanding of these intriguing enzymes.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, WI 53706, United States.