Repurposing drugs to target the malaria parasite unfolding protein response.
Chen, Y., Murillo-Solano, C., Kirkpatrick, M.G., Antoshchenko, T., Park, H.W., Pizarro, J.C.(2018) Sci Rep 8: 10333-10333
- PubMed: 29985421
- DOI: https://doi.org/10.1038/s41598-018-28608-2
- Primary Citation of Related Structures:
5UMB - PubMed Abstract:
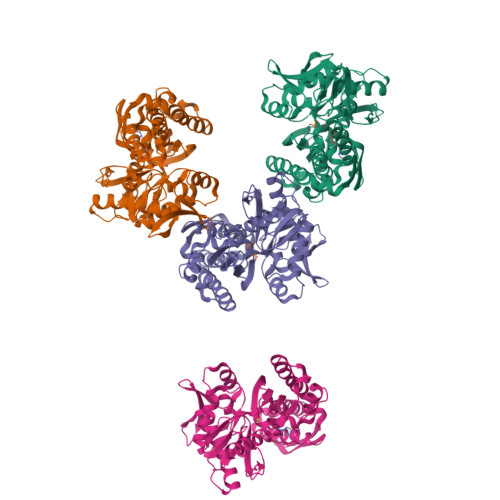
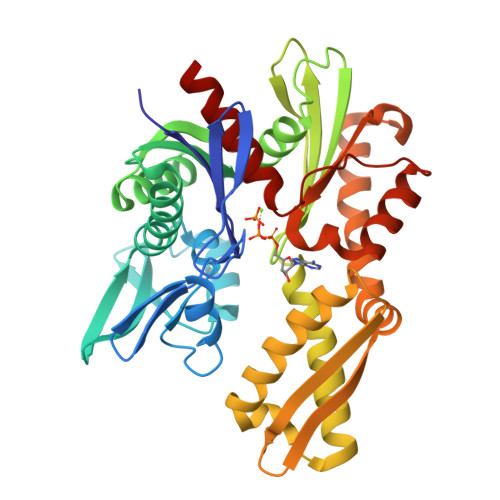
Drug resistant Plasmodium falciparum parasites represent a major obstacle in our efforts to control malaria, a deadly vector borne infectious disease. This situation creates an urgent need to find and validate new drug targets to contain the spread of the disease. Several genes associated with the unfolded protein response (UPR) including Glucose-regulated Protein 78 kDa (GRP78, also known as BiP) have been deemed potential drug targets. We explored the drug target potential of GRP78, a molecular chaperone that is a regulator of the UPR, for the treatment of P. falciparum parasite infection. By screening repurposed chaperone inhibitors that are anticancer agents, we showed that GRP78 inhibition is lethal to drug-sensitive and -resistant P. falciparum parasite strains in vitro. We correlated the antiplasmodial activity of the inhibitors with their ability to bind the malaria chaperone, by characterizing their binding to recombinant parasite GRP78. Furthermore, we determined the crystal structure of the ATP binding domain of P. falciparum GRP78 with ADP and identified structural features unique to the parasite. These data suggest that P. falciparum GRP78 can be a valid drug target and that its structural differences to human GRP78 emphasize potential to generate parasite specific compounds.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, School of Medicine, Tulane University, New Orleans, USA.