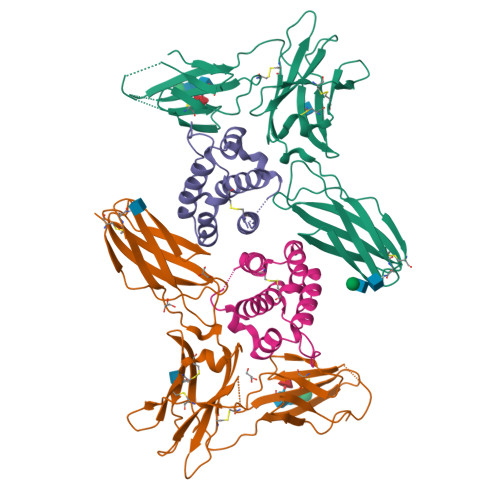
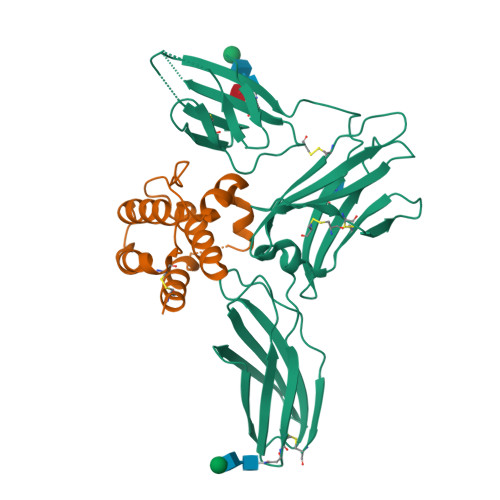
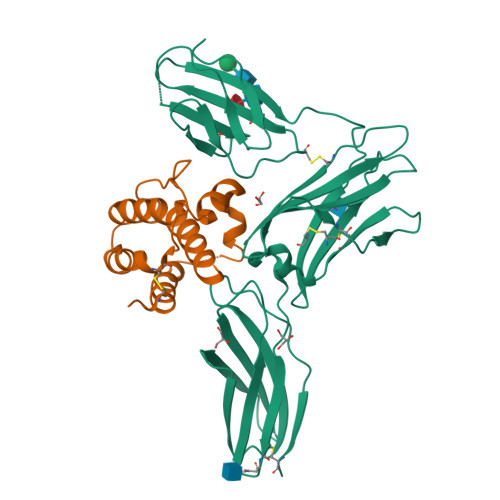
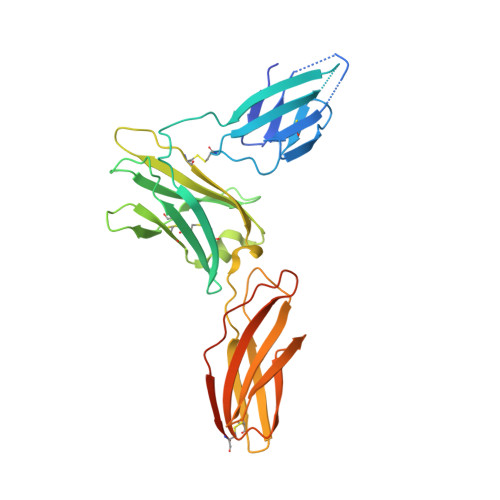
A dual role for the N-terminal domain of the IL-3 receptor in cell signalling.
Broughton, S.E., Hercus, T.R., Nero, T.L., Kan, W.L., Barry, E.F., Dottore, M., Cheung Tung Shing, K.S., Morton, C.J., Dhagat, U., Hardy, M.P., Wilson, N.J., Downton, M.T., Schieber, C., Hughes, T.P., Lopez, A.F., Parker, M.W.(2018) Nat Commun 9: 386-386
- PubMed: 29374162
- DOI: https://doi.org/10.1038/s41467-017-02633-7
- Primary Citation of Related Structures:
5UV8, 5UWC - PubMed Abstract:
The interleukin-3 (IL-3) receptor is a cell-surface heterodimer that links the haemopoietic, vascular and immune systems and is overexpressed in acute and chronic myeloid leukaemia progenitor cells. It belongs to the type I cytokine receptor family in which the α-subunits consist of two fibronectin III-like domains that bind cytokine, and a third, evolutionarily unrelated and topologically conserved, N-terminal domain (NTD) with unknown function. Here we show by crystallography that, while the NTD of IL3Rα is highly mobile in the presence of IL-3, it becomes surprisingly rigid in the presence of IL-3 K116W. Mutagenesis, biochemical and functional studies show that the NTD of IL3Rα regulates IL-3 binding and signalling and reveal an unexpected role in preventing spontaneous receptor dimerisation. Our work identifies a dual role for the NTD in this cytokine receptor family, protecting against inappropriate signalling and dynamically regulating cytokine receptor binding and function.
Organizational Affiliation:
Australian Cancer Research Foundation Rational Drug Discovery Centre, St. Vincent's Institute of Medical Research, Fitzroy, VIC, 3065, Australia.