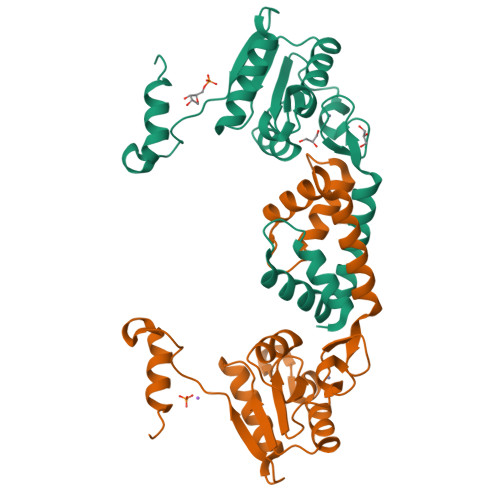
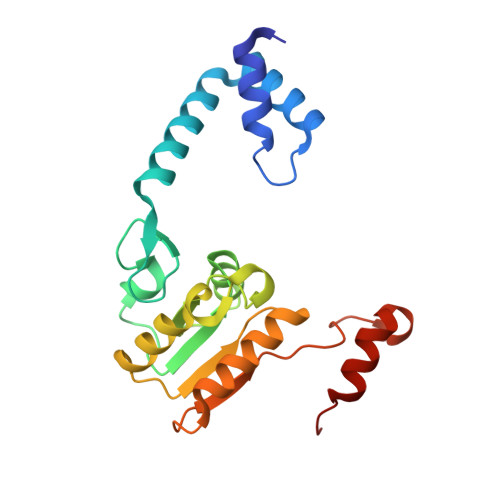
Crystal structure of RecR, a member of the RecFOR DNA-repair pathway, from Pseudomonas aeruginosa PAO1.
Che, S., Chen, Y., Liang, Y., Zhang, Q., Bartlam, M.(2018) Acta Crystallogr F Struct Biol Commun 74: 222-230
- PubMed: 29633970
- DOI: https://doi.org/10.1107/S2053230X18003503
- Primary Citation of Related Structures:
5Z2V - PubMed Abstract:
DNA damage is usually lethal to all organisms. Homologous recombination plays an important role in the DNA damage-repair process in prokaryotic organisms. Two pathways are responsible for homologous recombination in Pseudomonas aeruginosa: the RecBCD pathway and the RecFOR pathway. RecR is an important regulator in the RecFOR homologous recombination pathway in P. aeruginosa. It forms complexes with RecF and RecO that can facilitate the loading of RecA onto ssDNA in the RecFOR pathway. Here, the crystal structure of RecR from P. aeruginosa PAO1 (PaRecR) is reported. PaRecR crystallizes in space group P6 1 22, with two monomers per asymmetric unit. Analytical ultracentrifugation data show that PaRecR forms a stable dimer, but can exist as a tetramer in solution. The crystal structure shows that dimeric PaRecR forms a ring-like tetramer architecture via crystal symmetry. The presence of a ligand in the Walker B motif of one RecR subunit suggests a putative nucleotide-binding site.
Organizational Affiliation:
College of Life Sciences, Nankai University, 94 Weijin Road, Tianjin 300071, People's Republic of China.