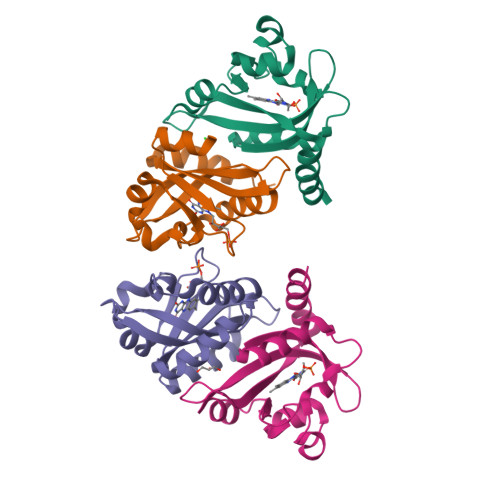
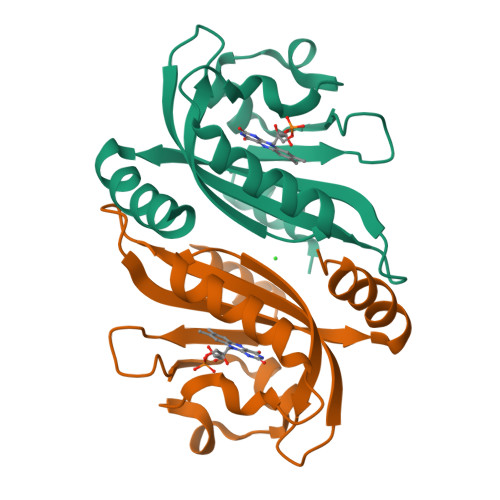
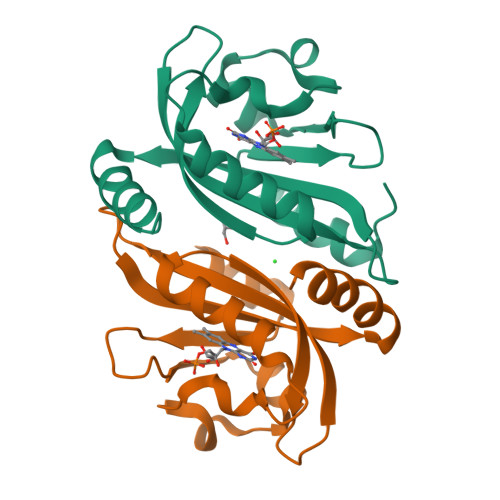
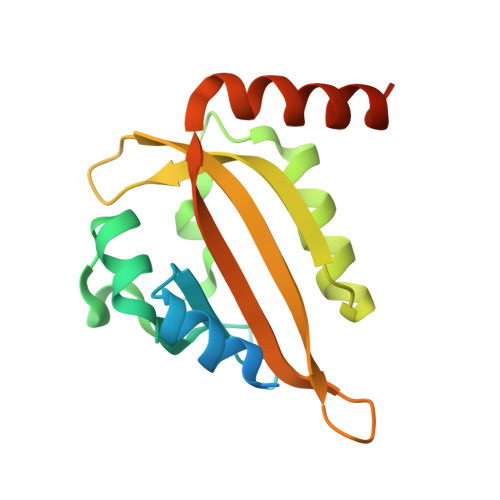
Structure of a Native-Like Aureochrome 1A Lov Domain Dimer from Phaeodactylum Tricornutum.
Banerjee, A., Herman, E., Kottke, T., Essen, L.O.(2016) Structure 24: 171
- PubMed: 26688213
- DOI: https://doi.org/10.1016/j.str.2015.10.022
- Primary Citation of Related Structures:
5A8B - PubMed Abstract:
Light-oxygen-voltage (LOV) domains absorb blue light for mediating various biological responses in all three domains of life. Aureochromes from stramenopile algae represent a subfamily of photoreceptors that differs by its inversed topology with a C-terminal LOV sensor and an N-terminal effector (basic region leucine zipper, bZIP) domain. We crystallized the LOV domain including its flanking helices, A'α and Jα, of aureochrome 1a from Phaeodactylum tricornutum in the dark state and solved the structure at 2.8 Å resolution. Both flanking helices contribute to the interface of the native-like dimer. Small-angle X-ray scattering shows light-induced conformational changes limited to the dimeric envelope as well as increased flexibility in the lit state for the flanking helices. These rearrangements are considered to be crucial for the formation of the light-activated dimer. Finally, the LOV domain of the class 2 aureochrome PtAUREO2 was shown to lack a chromophore because of steric hindrance caused by M301.
Organizational Affiliation:
Structural Biochemistry - Department of Chemistry, Philipps University Marburg, Hans-Meerwein Straße 4, 35032 Marburg, Germany.