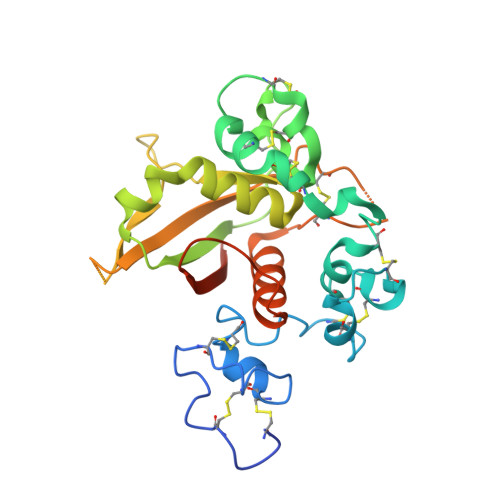
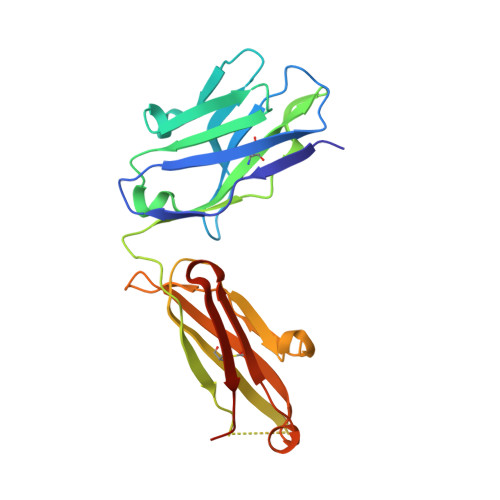
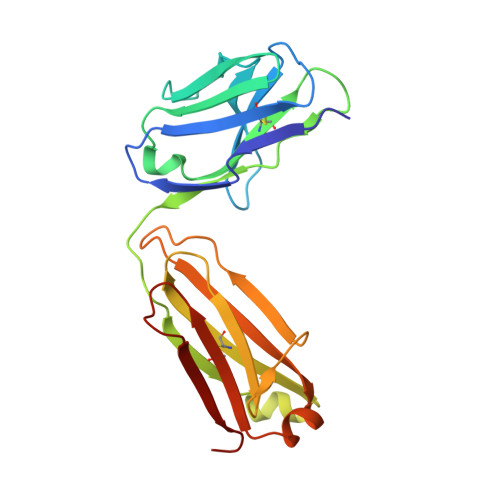
Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Bernasconi-Elias, P., Hu, T., Jenkins, D., Firestone, B., Gans, S., Kurth, E., Capodieci, P., Deplazes-Lauber, J., Petropoulos, K., Thiel, P., Ponsel, D., Hee Choi, S., LeMotte, P., London, A., Goetcshkes, M., Nolin, E., Jones, M.D., Slocum, K., Kluk, M.J., Weinstock, D.M., Christodoulou, A., Weinberg, O., Jaehrling, J., Ettenberg, S.A., Buckler, A., Blacklow, S.C., Aster, J.C., Fryer, C.J.(2016) Oncogene 35: 6077-6086
- PubMed: 27157619
- DOI: https://doi.org/10.1038/onc.2016.133
- Primary Citation of Related Structures:
5CZV, 5CZX - PubMed Abstract:
Notch receptors have been implicated as oncogenic drivers in several cancers, the most notable example being NOTCH1 in T-cell acute lymphoblastic leukemia (T-ALL). To characterize the role of activated NOTCH3 in cancer, we generated an antibody that detects the neo-epitope created upon gamma-secretase cleavage of NOTCH3 to release its intracellular domain (ICD3), and sequenced the negative regulatory region (NRR) and PEST (proline, glutamate, serine, threonine) domain coding regions of NOTCH3 in a panel of cell lines. We also characterize NOTCH3 tumor-associated mutations that result in activation of signaling and report new inhibitory antibodies. We determined the structural basis for receptor inhibition by obtaining the first co-crystal structure of a NOTCH3 antibody with the NRR protein and defined two distinct epitopes for NRR antibodies. The antibodies exhibit potent anti-leukemic activity in cell lines and tumor xenografts harboring NOTCH3 activating mutations. Screening of primary T-ALL samples reveals that 2 of 40 tumors examined show active NOTCH3 signaling. We also identified evidence of NOTCH3 activation in 12 of 24 patient-derived orthotopic xenograft models, 2 of which exhibit activation of NOTCH3 without activation of NOTCH1. Our studies provide additional insights into NOTCH3 activation and offer a path forward for identification of cancers that are likely to respond to therapy with NOTCH3 selective inhibitory antibodies.
Organizational Affiliation:
Developmental and Molecular Pathways, Novartis Institutes for Biomedical Research, Cambridge, MA, USA.