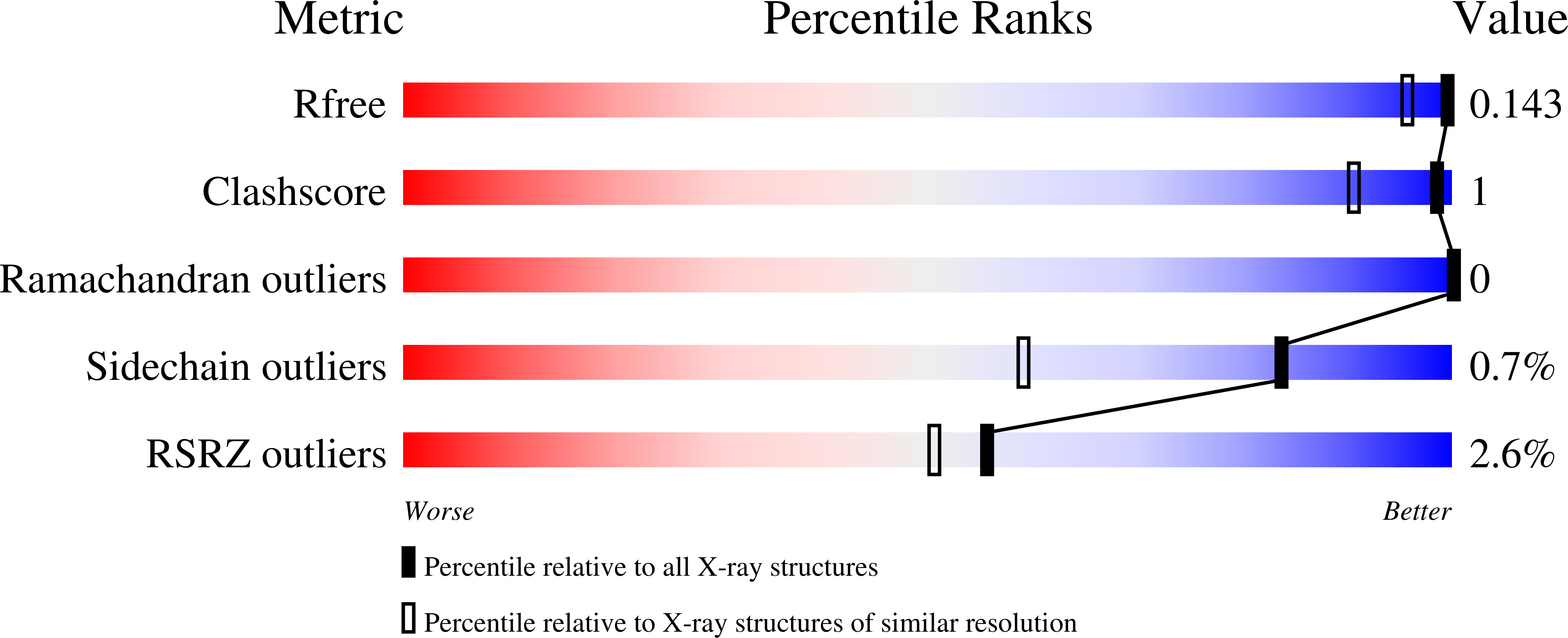
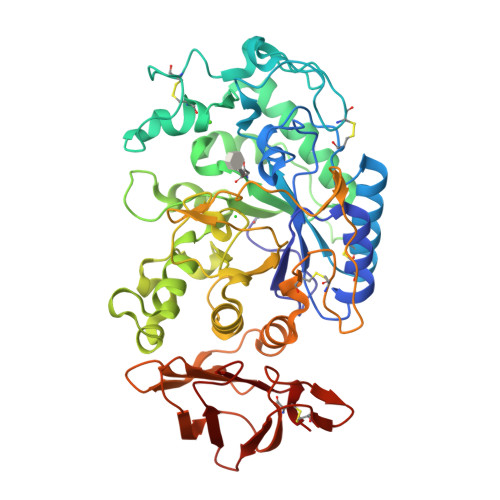
Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Caner, S., Zhang, X., Jiang, J., Chen, H.M., Nguyen, N.T., Overkleeft, H., Brayer, G.D., Withers, S.G.(2016) FEBS Lett 590: 1143-1151
- PubMed: 27000970
- DOI: https://doi.org/10.1002/1873-3468.12143
- Primary Citation of Related Structures:
5EMY - PubMed Abstract:
As part of a search for selective, mechanism-based covalent inhibitors of human pancreatic α-amylase we describe the chemoenzymatic synthesis of the disaccharide analog α-glucosyl epi-cyclophellitol, demonstrate its stoichiometric reaction with human pancreatic α-amylase and evaluate the time dependence of its inhibition. X-ray crystallographic analysis of the covalent derivative so formed confirms its reaction at the active site with formation of a covalent bond to the catalytic nucleophile D197. The structure illuminates the interactions with the active site and confirms OH4' on the nonreducing end sugar as a good site for attachment of fluorescent tags in generating probes for localization and quantitation of amylase in vivo.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of British Columbia, Vancouver, BC, Canada.