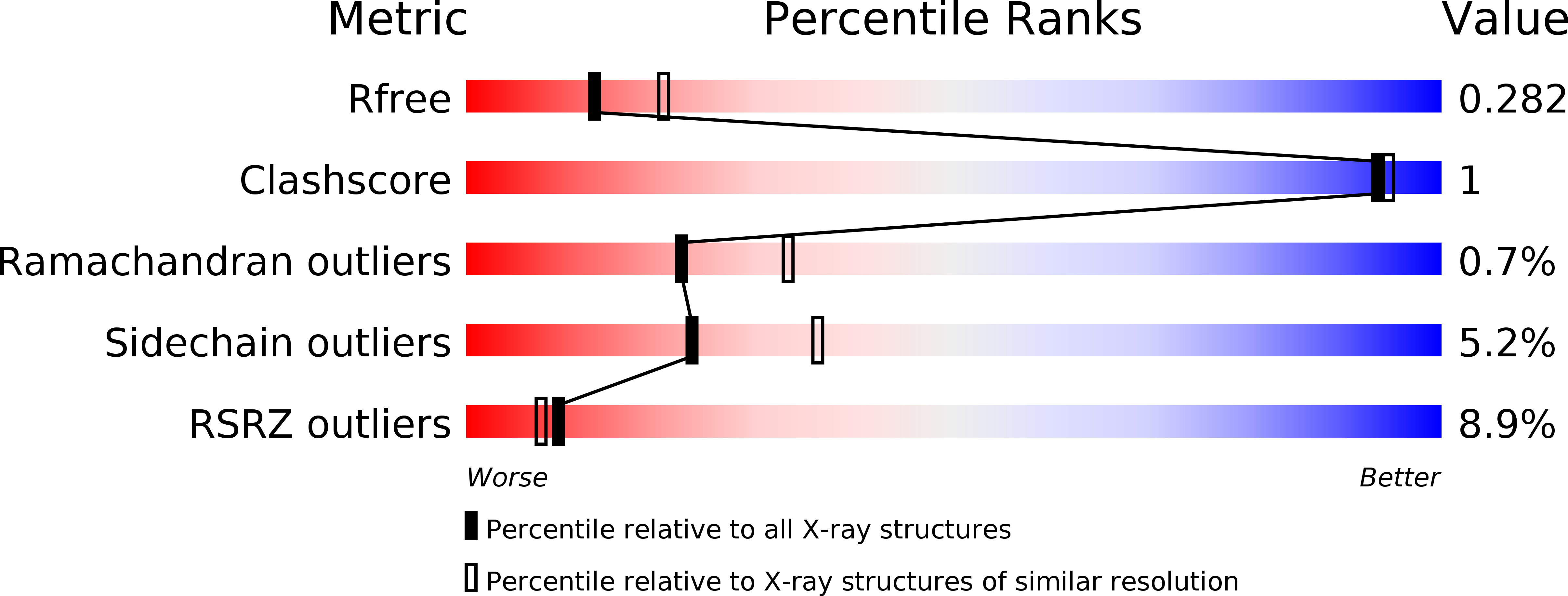
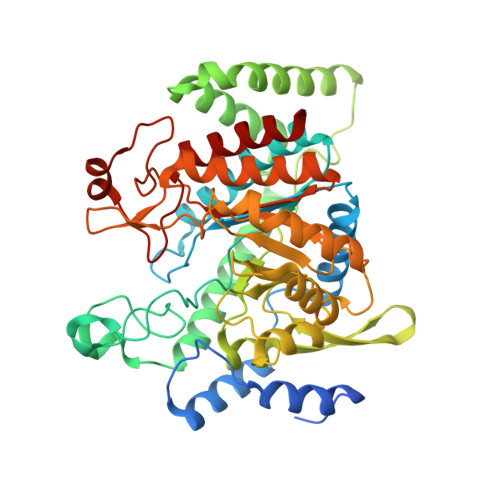
Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Burschowsky, D., Thorbjornsrud, H.V., Heim, J.B., Fahrig-Kamarauskaite, J.R., Wurth-Roderer, K., Kast, P., Krengel, U.(2018) Biochemistry 57: 557-573
- PubMed: 29178787
- DOI: https://doi.org/10.1021/acs.biochem.7b01018
- PubMed Abstract:
Corynebacterium glutamicum is widely used for the industrial production of amino acids, nucleotides, and vitamins. The shikimate pathway enzymes DAHP synthase (CgDS, Cg2391) and chorismate mutase (CgCM, Cgl0853) play a key role in the biosynthesis of aromatic compounds. Here we show that CgCM requires the formation of a complex with CgDS to achieve full activity, and that both CgCM and CgDS are feedback regulated by aromatic amino acids binding to CgDS. Kinetic analysis showed that Phe and Tyr inhibit CgCM activity by inter-enzyme allostery, whereas binding of Trp to CgDS strongly activates CgCM. Mechanistic insights were gained from crystal structures of the CgCM homodimer, tetrameric CgDS, and the heterooctameric CgCM-CgDS complex, refined to 1.1, 2.5, and 2.2 Å resolution, respectively. Structural details from the allosteric binding sites reveal that DAHP synthase is recruited as the dominant regulatory platform to control the shikimate pathway, similar to the corresponding enzyme complex from Mycobacterium tuberculosis.
Organizational Affiliation:
Department of Chemistry, University of Oslo , NO-0315 Oslo, Norway.