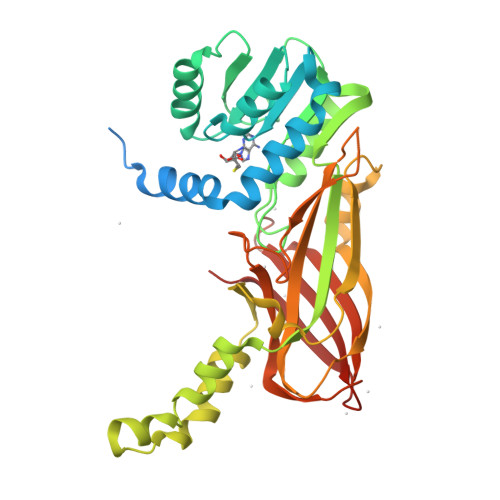
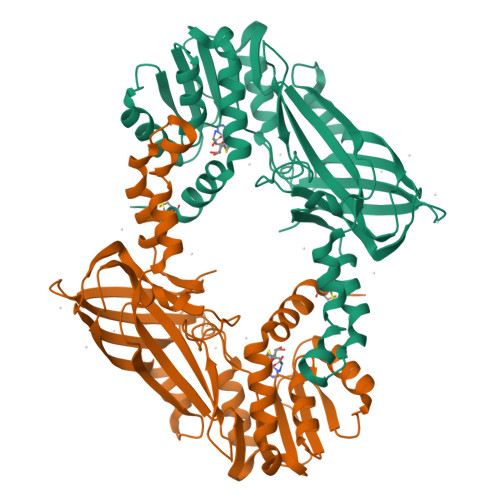
Structural basis of arginine asymmetrical dimethylation by PRMT6.
Wu, H., Zheng, W., Eram, M.S., Vhuiyan, M., Dong, A., Zeng, H., He, H., Brown, P., Frankel, A., Vedadi, M., Luo, M., Min, J.(2016) Biochem J 473: 3049-3063
- PubMed: 27480107
- DOI: https://doi.org/10.1042/BCJ20160537
- Primary Citation of Related Structures:
4HC4, 4QQK, 5HZM - PubMed Abstract:
PRMT6 is a type I protein arginine methyltransferase, generating the asymmetric dimethylarginine mark on proteins such as histone H3R2. Asymmetric dimethylation of histone H3R2 by PRMT6 acts as a repressive mark that antagonizes trimethylation of H3 lysine 4 by the MLL histone H3K4 methyltransferase. PRMT6 is overexpressed in several cancer types, including prostate, bladder and lung cancers; therefore, it is of great interest to develop potent and selective inhibitors for PRMT6. Here, we report the synthesis of a potent bisubstrate inhibitor GMS [6'-methyleneamine sinefungin, an analog of sinefungin (SNF)], and the crystal structures of human PRMT6 in complex, respectively, with S-adenosyl-L-homocysteine (SAH) and the bisubstrate inhibitor GMS that shed light on the significantly improved inhibition effect of GMS on methylation activity of PRMT6 compared with SAH and an S-adenosyl-L-methionine competitive methyltransferase inhibitor SNF. In addition, we also crystallized PRMT6 in complex with SAH and a short arginine-containing peptide. Based on the structural information here and available in the PDB database, we proposed a mechanism that can rationalize the distinctive arginine methylation product specificity of different types of arginine methyltransferases and pinpoint the structural determinant of such a specificity.
Organizational Affiliation:
Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5J 1L7, Canada.