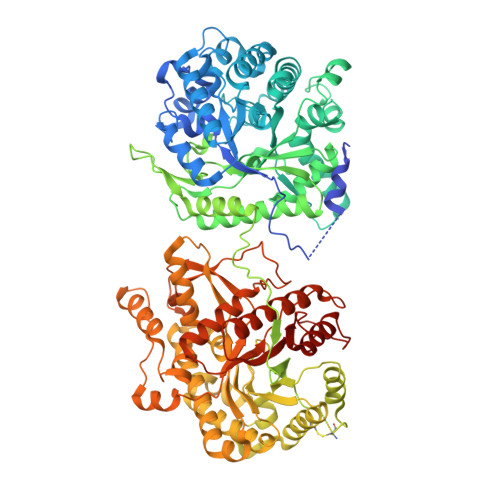
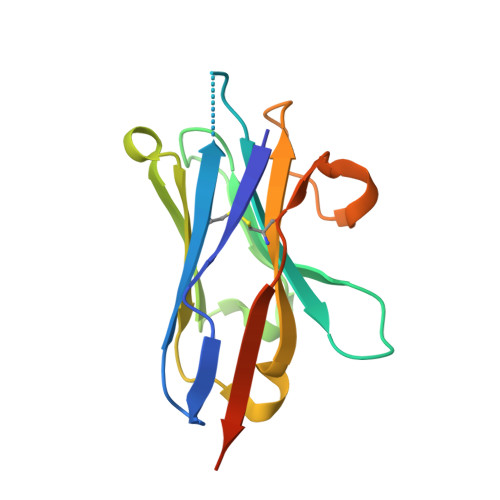
Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Lee, S., Choi, J., Mohanty, J., Sousa, L.P., Tome, F., Pardon, E., Steyaert, J., Lemmon, M.A., Lax, I., Schlessinger, J.(2018) Nature 553: 501-505
- PubMed: 29342135
- DOI: https://doi.org/10.1038/nature25010
- Primary Citation of Related Structures:
5VAK, 5VAN, 5VAQ - PubMed Abstract:
Canonical fibroblast growth factors (FGFs) activate FGF receptors (FGFRs) through paracrine or autocrine mechanisms in a process that requires cooperation with heparan sulfate proteoglycans, which function as co-receptors for FGFR activation. By contrast, endocrine FGFs (FGF19, FGF21 and FGF23) are circulating hormones that regulate critical metabolic processes in a variety of tissues. FGF19 regulates bile acid synthesis and lipogenesis, whereas FGF21 stimulates insulin sensitivity, energy expenditure and weight loss. Endocrine FGFs signal through FGFRs in a manner that requires klothos, which are cell-surface proteins that possess tandem glycosidase domains. Here we describe the crystal structures of free and ligand-bound β-klotho extracellular regions that reveal the molecular mechanism that underlies the specificity of FGF21 towards β-klotho and demonstrate how the FGFR is activated in a klotho-dependent manner. β-Klotho serves as a primary 'zip code'-like receptor that acts as a targeting signal for FGF21, and FGFR functions as a catalytic subunit that mediates intracellular signalling. Our structures also show how the sugar-cutting enzyme glycosidase has evolved to become a specific receptor for hormones that regulate metabolic processes, including the lowering of blood sugar levels. Finally, we describe an agonistic variant of FGF21 with enhanced biological activity and present structural insights into the potential development of therapeutic agents for diseases linked to endocrine FGFs.
Organizational Affiliation:
Department of Pharmacology and Yale Cancer Biology Institute, Yale School of Medicine, 333 Cedar Street, New Haven, Connecticut 06520, USA.