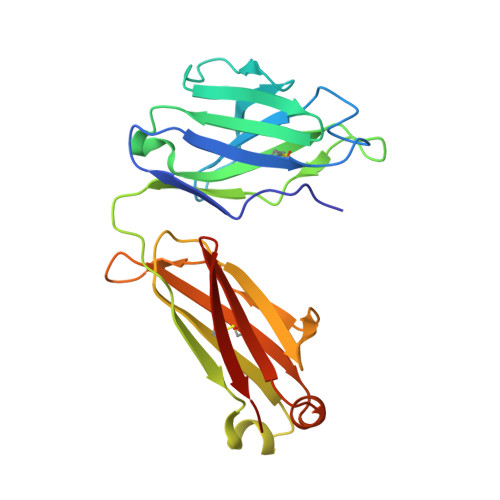
Structural basis for antibody-mediated neutralization of Lassa virus.
Hastie, K.M., Zandonatti, M.A., Kleinfelter, L.M., Heinrich, M.L., Rowland, M.M., Chandran, K., Branco, L.M., Robinson, J.E., Garry, R.F., Saphire, E.O.(2017) Science 356: 923-928
- PubMed: 28572385
- DOI: https://doi.org/10.1126/science.aam7260
- Primary Citation of Related Structures:
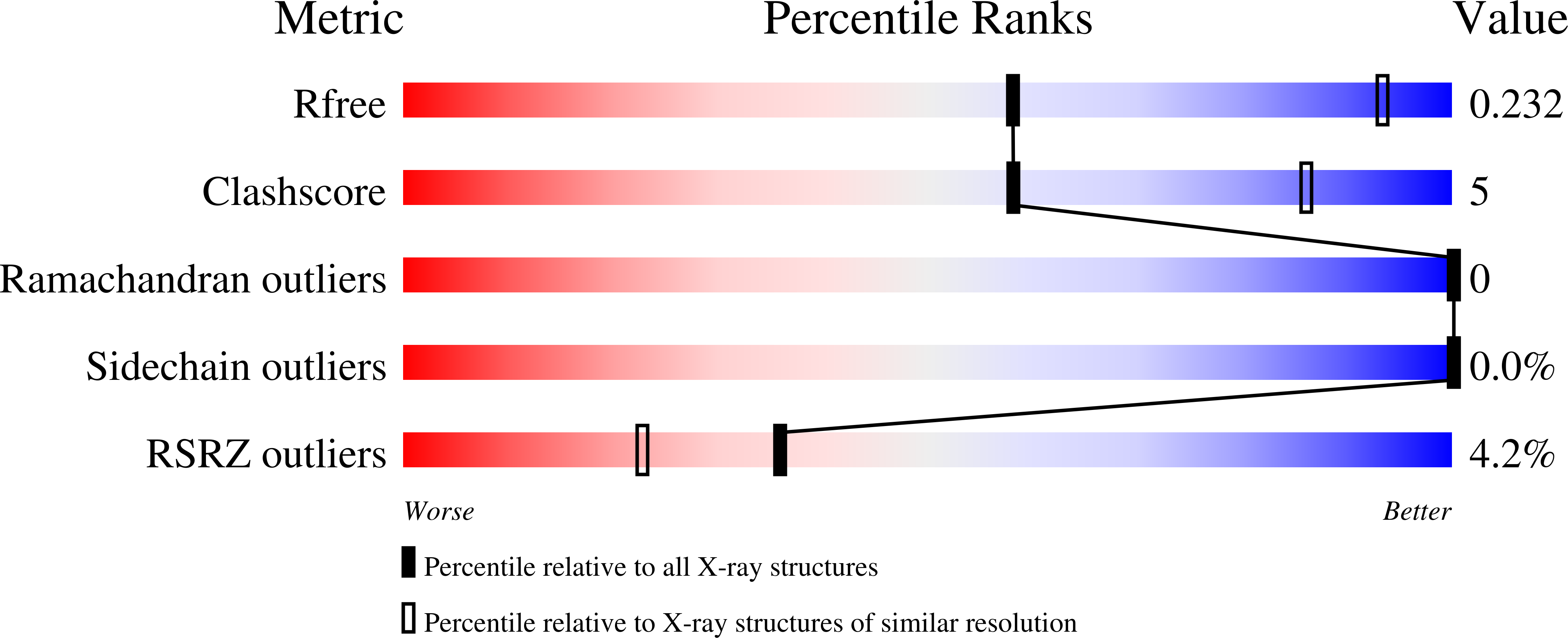
5VK2 - PubMed Abstract:
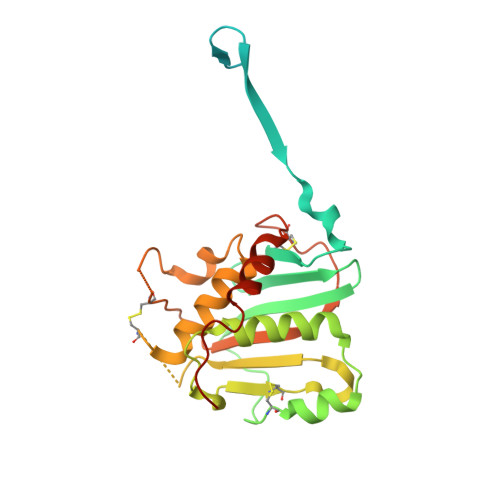
The arenavirus Lassa causes severe hemorrhagic fever and a significant disease burden in West Africa every year. The glycoprotein, GPC, is the sole antigen expressed on the viral surface and the critical target for antibody-mediated neutralization. Here we present the crystal structure of the trimeric, prefusion ectodomain of Lassa GP bound to a neutralizing antibody from a human survivor at 3.2-angstrom resolution. The antibody extensively anchors two monomers together at the base of the trimer, and biochemical analysis suggests that it neutralizes by inhibiting conformational changes required for entry. This work illuminates pH-driven conformational changes in both receptor-binding and fusion subunits of Lassa virus, illustrates the unique assembly of the arenavirus glycoprotein spike, and provides a much-needed template for vaccine design against these threats to global health.
Organizational Affiliation:
Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA 92037, USA.