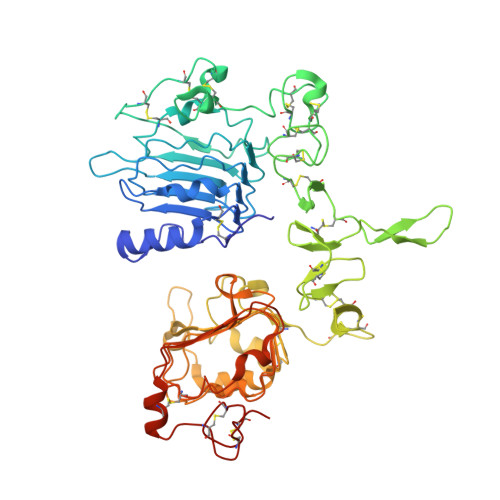
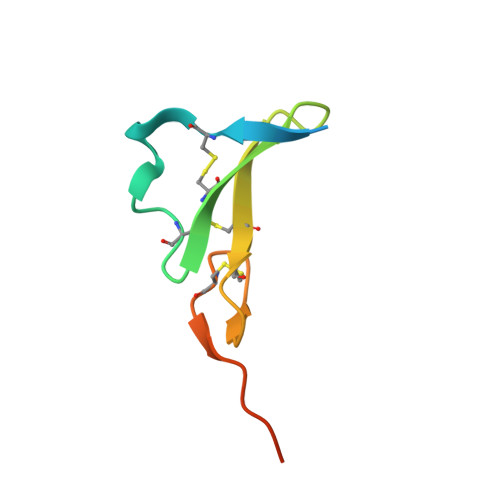
EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Freed, D.M., Bessman, N.J., Kiyatkin, A., Salazar-Cavazos, E., Byrne, P.O., Moore, J.O., Valley, C.C., Ferguson, K.M., Leahy, D.J., Lidke, D.S., Lemmon, M.A.(2017) Cell 171: 683-695.e18
- PubMed: 28988771
- DOI: https://doi.org/10.1016/j.cell.2017.09.017
- Primary Citation of Related Structures:
5WB7, 5WB8 - PubMed Abstract:
Epidermal growth factor receptor (EGFR) regulates many crucial cellular programs, with seven different activating ligands shaping cell signaling in distinct ways. Using crystallography and other approaches, we show how the EGFR ligands epiregulin (EREG) and epigen (EPGN) stabilize different dimeric conformations of the EGFR extracellular region. As a consequence, EREG or EPGN induce less stable EGFR dimers than EGF-making them partial agonists of EGFR dimerization. Unexpectedly, this weakened dimerization elicits more sustained EGFR signaling than seen with EGF, provoking responses in breast cancer cells associated with differentiation rather than proliferation. Our results reveal how responses to different EGFR ligands are defined by receptor dimerization strength and signaling dynamics. These findings have broad implications for understanding receptor tyrosine kinase (RTK) signaling specificity. Our results also suggest parallels between partial and/or biased agonism in RTKs and G-protein-coupled receptors, as well as new therapeutic opportunities for correcting RTK signaling output.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520, USA; Yale Cancer Biology Institute, Yale University, West Haven, CT 06516, USA.