Phosphorylated and non-phosphorylated HCK kinase domains produced by cell-free protein expression.
Katsura, K., Tomabechi, Y., Matsuda, T., Yonemochi, M., Mikuni, J., Ohsawa, N., Terada, T., Yokoyama, S., Kukimoto-Niino, M., Takemoto, C., Shirouzu, M.(2018) Protein Expr Purif 150: 92-99
- PubMed: 29793032
- DOI: https://doi.org/10.1016/j.pep.2018.05.005
- Primary Citation of Related Structures:
5ZJ6 - PubMed Abstract:
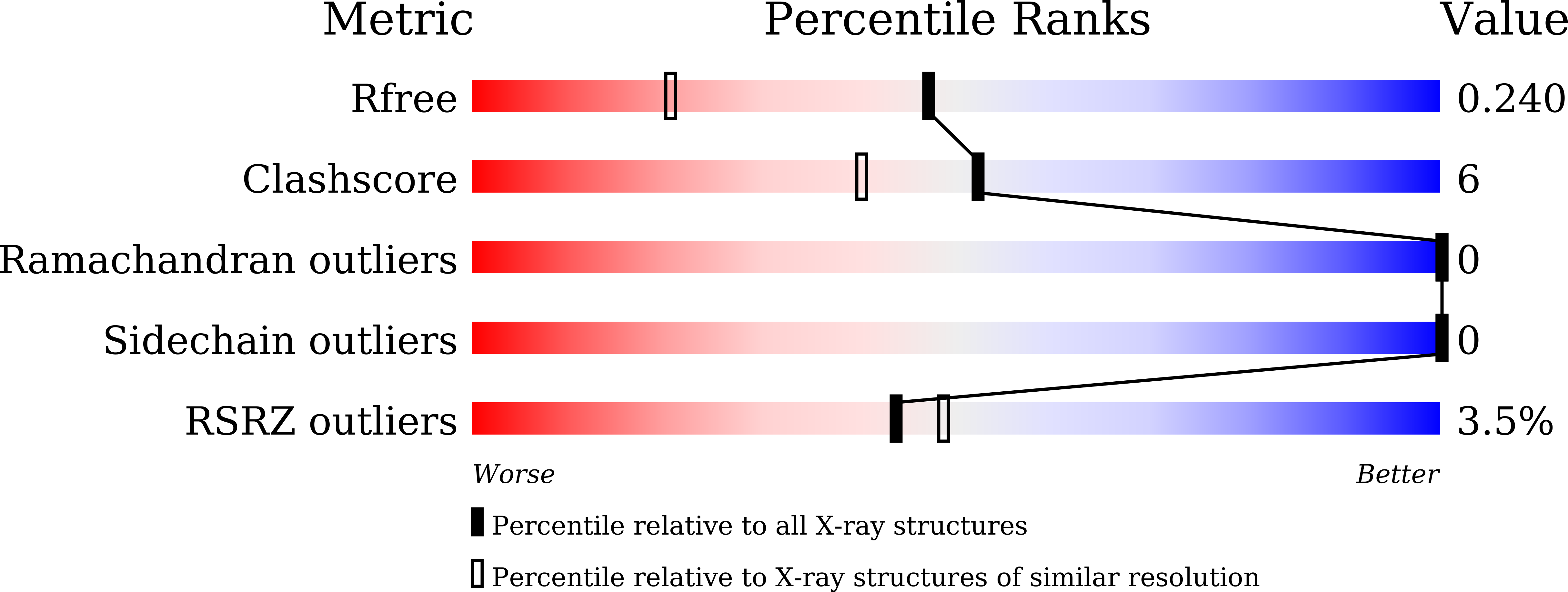
Since phosphorylation is involved in various physiological events, kinases and interacting factors can be potential targets for drug discovery. For the development and improvement of inhibitors from the point of view of mechanistic enzymology, a cell-free protein synthesis system would be advantageous, since it could prepare mutant proteins easily. However, especially in the case of protein kinase, product solubility remains one of the major challenges. To overcome this problem, we prepared a chaperone-supplemented extract from Escherichia coli BL21 cells harboring a plasmid encoding a set of chaperone genes, dnaK, dnaJ, and grpE. We explored cell-disruption procedures and constructed an efficient protein synthesis system. Employing this system, we produced the kinase domain of human hematopoietic cell kinase (HCK) to obtain further structural information about its molecular interaction with one of its inhibitors, previously developed by our group (RK-20449). Lower reaction temperature improved the solubility, and addition of a protein phosphatase (YpoH) facilitated the homogeneous production of the non-phosphorylated kinase domain. Crystals of the purified product were obtained and the kinase-inhibitor complex structure was solved at 1.7 Å resolution. In addition, results of kinase activity measurement, using a synthetic substrate, showed that the kinase activity was facilitated by autophosphorylation at Tyr416, as confirmed by the peptide mass mapping.
Organizational Affiliation:
Protein Functional and Structural Biology Team, RIKEN Center for Life Science Technology, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.